20.109(S21):M1D1
Introduction:In silico cloning library
Restriction enzyme digest
Restriction endonucleases, also called restriction enzymes, 'cut' or 'digest' DNA at specific sequences of bases. The restriction enzymes are named according to the prokaryotic organism from which they were isolated. For example, the restriction endonuclease EcoRI (pronounced “echo-are-one”) was originally isolated from E. coli giving it the “Eco” part of the name. “RI” indicates the particular version on the E. coli strain (RY13) and the fact that it was the first restriction enzyme isolated from this strain.
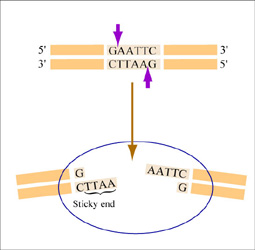
The sequence of DNA that is bound and cleaved by an endonuclease is called the recognition sequence or restriction site. These sequences are usually four or six base pairs long and palindromic, that is, they read the same 5’ to 3’ on the top and bottom strand of DNA. For example, the recognition sequence for EcoRI is 5’ GAATTC 3’ (see figure at right). EcoRI cleaves the phosphate backbone of DNA between the G and A of the recognition sequence, which generates overhangs or 'sticky ends' of double-stranded DNA.
Unlike EcoRI, some other restriction enzymes cut precisely in the middle of the palindromic DNA sequence, thus leaving no overhangs after digestion. The single-stranded overhangs resulting from DNA digestion by enzymes such as EcoRI are called sticky ends, while double-stranded ends resulting from digestion by enzymes such as HaeIII are called blunt ends. HaeIII recognizes 5’ GGCC 3’ and upon recognition cuts in the center of the sequence.