Difference between revisions of "20.109(S21):M1D1"
(→Part 2: Mutagenesis of scFv clone using Error-prone PCR) |
(→Part 2: Mutagenesis of scFv clone using Error-prone PCR) |
||
| Line 31: | Line 31: | ||
Based on the numerous applications of PCR, it may seem that the technique has been around forever. In fact it is just under 30 years old. In 1984, Kary Mullis described this technique for amplifying DNA of known or unknown sequence, realizing immediately the significance of his insight. ''"Dear Thor!," I exclaimed. I had solved the most annoying problems in DNA chemistry in a single lightening bolt. Abundance and distinction. With two oligonucleotides, DNA polymerase, and the four nucleosidetriphosphates I could make as much of a DNA sequence as I wanted and I could make it on a fragment of a specific size that I could distinguish easily. Somehow, I thought, it had to be an illusion. Otherwise it would change DNA chemistry forever. Otherwise it would make me famous. It was too easy. Someone else would have done it and I would surely have heard of it. We would be doing it all the time. What was I failing to see? "Jennifer, wake up. I've thought of something incredible." '' --Kary Mullis from his Nobel lecture; December 8, 1993 | Based on the numerous applications of PCR, it may seem that the technique has been around forever. In fact it is just under 30 years old. In 1984, Kary Mullis described this technique for amplifying DNA of known or unknown sequence, realizing immediately the significance of his insight. ''"Dear Thor!," I exclaimed. I had solved the most annoying problems in DNA chemistry in a single lightening bolt. Abundance and distinction. With two oligonucleotides, DNA polymerase, and the four nucleosidetriphosphates I could make as much of a DNA sequence as I wanted and I could make it on a fragment of a specific size that I could distinguish easily. Somehow, I thought, it had to be an illusion. Otherwise it would change DNA chemistry forever. Otherwise it would make me famous. It was too easy. Someone else would have done it and I would surely have heard of it. We would be doing it all the time. What was I failing to see? "Jennifer, wake up. I've thought of something incredible." '' --Kary Mullis from his Nobel lecture; December 8, 1993 | ||
| − | Please watch this animation of PCR created by LabXchange: [[https://www.labxchange.org/library/items/lb:LabXchange:4fdc8a5c:video:1 PCR Animation]]This will help answer questions for your lab notebook. | + | '''Please watch this animation of PCR created by LabXchange:''' [[https://www.labxchange.org/library/items/lb:LabXchange:4fdc8a5c:video:1 PCR Animation]]This will help answer questions for your lab notebook. |
<font color = #4a9152 >'''In your laboratory notebook,'''</font color> complete the following: | <font color = #4a9152 >'''In your laboratory notebook,'''</font color> complete the following: | ||
Revision as of 15:32, 1 February 2021
Contents
- 1 Introduction: Generate single-chain antibody fragment (scFv) library
- 2 Protocols:
- 2.1 Part 1: Investigate our parental scFv clone: plasmid ADI-31375
- 2.2 Part 2: Mutagenesis of scFv clone using Error-prone PCR
- 2.3 Part 3: Gel purify PCR products
- 2.4 Part 4: Prepare error-prone PCR products and backbone for yeast transformation
- 2.5 Part 5: Transform DNA into yeast
- 2.6 Part 6: Complete Orientation quiz
- 3 Navigation links
Introduction: Generate single-chain antibody fragment (scFv) library
To engineer a protein-protein interaction (antibody-antigen), we will use a method called directed evolution. Directed evolution is defined as an iterative process of mutating a gene, then screening those mutants for a particular functionality, collecting the best mutants and repeating. It is a method that is inspired by the process of natural selection.
In the Wittrup lab protocol, successive rounds of random mutagenesis of the scFv gene, followed by expression of the scFv on the surface of yeast then selection for "good binders" by fluorescence activated cell sorting is carried out. This process will produce an antibody fragment with increased affinity or stability to its antigen.
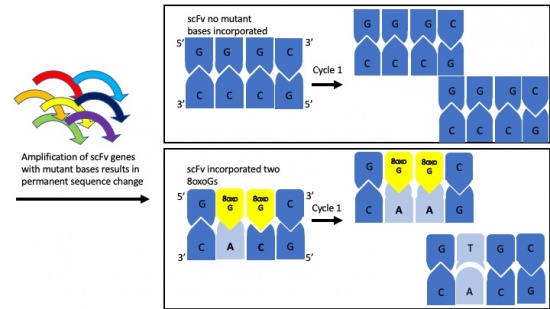
Today we will walk through the technical details of random DNA mutagenesis via error-prone PCR. This process creates a pool of scFv genes that have random mutations throughout the DNA sequence. This collection of varied scFV genes are called a library.
Finally, we will outline how one integrates this library into an expression vector, also referred to as the backbone or plasmid, that can then be transformed into yeast and expressed on the surface of the cells. This method is called cloning. Integral tools when cloning DNA plasmids are restriction endonucleases.
Protocols:
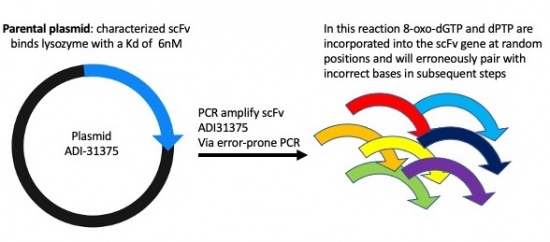
Part 1: Investigate our parental scFv clone: plasmid ADI-31375
Motivation: To understand the important features of scFv yeast display it's important to know what genes are being utilized to control this system.
We will use SnapGene software for our DNA visualization and alignments. Feel free to use any software you are familiar with to carry out analysis. If you are off campus you must log into a VPN connection prior to opening the SnapGene application. Here is the link to the VPN download and installation instructions. You may need to update the MIT SnapGene license number if you have not opened the application recently. The new license information can be found here.
Part 2: Mutagenesis of scFv clone using Error-prone PCR
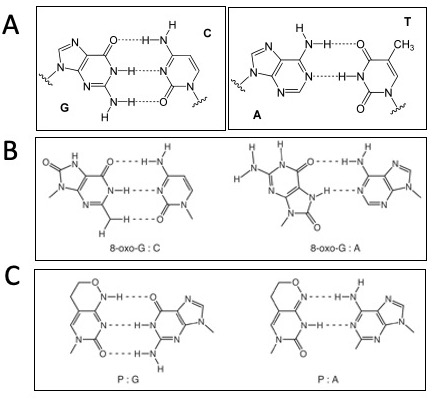
Motivation: To introduce random DNA mutation into our scFv clone we will carry out error-prone polymerase chain reaction (PCR). We will use special bases in the PCR reaction that mimic DNA damage and can erroneously pair with incorrect bases and will result in changes in the scFv DNA sequence and when expressed in yeast, changes to the scFv protein sequence.
PCR amplification The applications of PCR (polymerase chain reaction) are widespread, from forensics to molecular biology to evolution, but the goal of any PCR is the same: to generate many copies of DNA from a single or a few specific sequence(s) (called the “target” or “template”).
In addition to the target, PCR requires only three components: primers to bind sequence flanking the target, dNTPs to polymerize, and a heat-stable polymerase to carry out the synthesis reaction over and over and over. DNA polymerases require short initating pieces of DNA (or RNA) called primers in order to copy DNA. In PCR amplification, forward and reverse primers that target the non-coding and coding strands of DNA, respectively, are separated by a distance equal to the length of the DNA to be copied. PCR is a three-step process (denature, anneal, extend) and these steps are repeated. After 30 cycles of PCR, there could be as many as a billion copies of the original target sequence.
Based on the numerous applications of PCR, it may seem that the technique has been around forever. In fact it is just under 30 years old. In 1984, Kary Mullis described this technique for amplifying DNA of known or unknown sequence, realizing immediately the significance of his insight. "Dear Thor!," I exclaimed. I had solved the most annoying problems in DNA chemistry in a single lightening bolt. Abundance and distinction. With two oligonucleotides, DNA polymerase, and the four nucleosidetriphosphates I could make as much of a DNA sequence as I wanted and I could make it on a fragment of a specific size that I could distinguish easily. Somehow, I thought, it had to be an illusion. Otherwise it would change DNA chemistry forever. Otherwise it would make me famous. It was too easy. Someone else would have done it and I would surely have heard of it. We would be doing it all the time. What was I failing to see? "Jennifer, wake up. I've thought of something incredible." --Kary Mullis from his Nobel lecture; December 8, 1993
Please watch this animation of PCR created by LabXchange: [PCR Animation]This will help answer questions for your lab notebook.
In your laboratory notebook, complete the following:
| Component, concentration | Final concentration | Volume |
|---|---|---|
| 10X Taq buffer | 1X | |
| 50mM MgCl2 | 2mM | |
| 10uM Forward primer | 2mM | |
| 10uM Reverse primer | 2mM | |
| 10mM dNTPs | 200uM | |
| 100pg/uL template DNA | 13.3 pg/uL | |
| 20uM 8-oxo-dGTP | 2uM | |
| 20uM dPTP | 2uM | |
| 5 units/uL Taq polymerase | 0.05 units/uL | |
| dH2O |
- Retrieve a 200uL thin-walled PCR tube and combine the components above.
- It is best practice to add water first and enzyme (Taq) last.
- The template DNA is our parental scFv clone, ADI-31375.
- Optimal primers have been determined experimentally by the Wittrup lab.
- Forward 5'-CGACGATTGAAGGTAGATACCCATACGACGTTCCAGACTACGCTCTGCAG-3'
- Reverse 5'-CAGATCTCGAGCTATTACAAGTCCTCTTCAGAAATAAGCTTTTGTTC-3'
- Amplify DNA on the thermocycler using the following program:
| Cycles | Denaturation | Annealing | Polymerization |
|---|---|---|---|
| 1 | 94°C for 3m | ||
| 2-11 | 94°C for 45s | 60°C for 30s | 72°C for 90s |
| 12 | 72°C for 10m |
Part 3: Gel purify PCR products
Motivation: To separate the PCR products from the template DNA we will use gel electrophoresis. We can use gel purification to exploit the difference in size between the DNA template and PCR product.
Electrophoresis is a technique that separates large molecules by size using an applied electrical field and a sieving matrix. DNA, RNA and proteins are the molecules most often studied with this technique; agarose and acrylamide gels are the two most common sieves. The molecules to be separated enter the matrix through a well at one end and are pulled through the matrix when a current is applied across it. The larger molecules get entwined in the matrix and are stalled; the smaller molecules wind through the matrix more easily and travel farther away from the well. The distance a DNA fragment travels is inversely proportional to the log of its length. Over time fragments of similar length accumulate into “bands” in the gel. Higher concentrations of agarose can be used to resolve smaller DNA fragments.
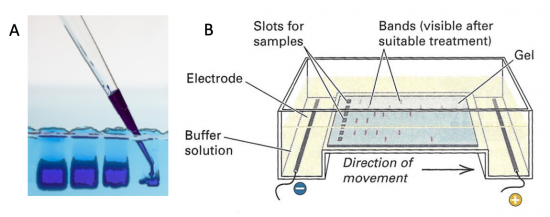
DNA and RNA are negatively charged molecules due to their phosphate backbone, and they naturally travel toward the positive electrode at the far end of the gel. Today you will separate DNA fragments using an agarose matrix. Agarose is a polymer that comes from seaweed. To prepare these gels, agarose and 1X TAE buffer (Tris base, acetic acid, and EDTA) are microwaved until the agarose is melted and fully dissolved. The molten agar is then poured into a horizontal casting tray, and a comb is added. Once the agar has solidified, the comb is removed, leaving wells into which the DNA samples can be loaded.
To purify the PCR mutant product from the parental template, a 1% agarose gel with SYBR Safe DNA stain was used to separate the DNA fragments. In addition, a well was loaded with a molecular weight marker (also called a DNA ladder) to determine the size of the fragments.
To ensure the steps included below are clear, please watch the video tutorial linked here: [DNA gel electrophoresis]. The steps are detailed below so you can follow along!
- Add 5 μL of 6x loading dye to the PCR product mix.
- Loading dye contains bromophenol blue as a tracking dye, which enables you to follow the progress of the electrophoresis.
- Glycerol is also included to weight the samples such that the liquid sinks into well.
- Flick the eppendorf tubes to mix the contents, then quick spin them in the microfuge to bring the contents of the tubes to the bottom.
- Load 25 μL of the reactant into the gel, as well as 10 μL of 1kb DNA ladder.
- Be sure to record the order in which you load your samples!
- To load your samples
Part 4: Prepare error-prone PCR products and backbone for yeast transformation
Motivation: To transform the mutated scFv products and the backbone in yeast we must carry out two additional steps. First we will amplify the PCR products so there is a sufficient amount of DNA for transformation nd second we will digest the backbone plasmid so the new mutated scFv genes can integrate into the plasmid.
| Component, concentration | Final concentration | Volume |
|---|---|---|
| 10X Taq buffer | 1X | 10uL |
| 50mM MgCl2 | 2mM | 4uL |
| 10uM Forward primer | 2mM | 5uL |
| 10uM Reverse primer | 2mM | 5uL |
| 10mM dNTPs | 200uM | 2.0uL |
| Gel purified PCR product | 4uL | |
| 5 units/uL Taq polymerase | 0.05 units/uL | 1uL |
| dH2O | 69uL |
| Cycles | Denaturation | Annealing | Polymerization |
|---|---|---|---|
| 1 | 94°C for 3m | ||
| 2-31 | 94°C for 45s | 60°C for 30s | 72°C for 90s |
| 32 | 72°C for 10m |
Restriction enzyme digests
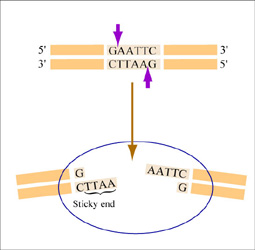
Restriction endonucleases, also called restriction enzymes, 'cut' or 'digest' DNA at specific sequences of bases. The restriction enzymes are named according to the prokaryotic organism from which they were isolated. For example, the restriction endonuclease EcoRI (pronounced “echo-are-one”) was originally isolated from E. coli giving it the “Eco” part of the name. “RI” indicates the particular version on the E. coli strain (RY13) and the fact that it was the first restriction enzyme isolated from this strain.
The sequence of DNA that is bound and cleaved by an endonuclease is called the recognition sequence or restriction site. These sequences are usually four or six base pairs long and palindromic, that is, they read the same 5’ to 3’ on the top and bottom strand of DNA. For example, the recognition sequence for EcoRI is 5’ GAATTC 3’ (see figure at right). EcoRI cleaves the phosphate backbone of DNA between the G and A of the recognition sequence, which generates overhangs or 'sticky ends' of double-stranded DNA.
Unlike EcoRI, some other restriction enzymes cut precisely in the middle of the palindromic DNA sequence, thus leaving no overhangs after digestion. The single-stranded overhangs resulting from DNA digestion by enzymes such as EcoRI are called sticky ends, while double-stranded ends resulting from digestion by enzymes such as HaeIII are called blunt ends. HaeIII recognizes 5’ GGCC 3’ and upon recognition cuts in the center of the sequence.
Part 5: Transform DNA into yeast
Motivation: To express the new scFv library we must transform the DNA into the yeast cells and select for circularized plasmid DNA.
We will use a transformation procedure called electroporation to get the DNA inside yeast cells. During this procedure yeast, specialized salt buffers, and DNA are loaded into a metal cuvette and a high voltage electrical pulse is used to open pores within the cell membrane. DNA is taken into the yeast cells and the pores close within minutes as the cells recover in full nutrient media.
Part 6: Complete Orientation quiz
Complete the orientation quiz with your partner. Though you are working with your partner, each student should record all answers independently. If you disagree with your partner on an answer, you should write what you think is the correct answer on your quiz.
Good luck!
Next day: Enrich candidate clones from library using FACS