Difference between revisions of "20.109(S23):M2D6"
Becky Meyer (Talk | contribs) (→Part 2: Perform metal uptake experiment) |
Becky Meyer (Talk | contribs) (→Part 2: Perform metal uptake experiment) |
||
| Line 26: | Line 26: | ||
**Use 1ml SD-G for a blank. | **Use 1ml SD-G for a blank. | ||
**Record these numbers in your notebook | **Record these numbers in your notebook | ||
| − | *Using SD-G media as a diluent, dilute each of your cultures to | + | *Using SD-G media as a diluent, dilute each of your cultures to OD<sub>600</sub> ~ 1.0 and a final volume of 8ml. This does not have to be exact, but the cultures should have a similar OD<sub>600</sub> before you begin the experiment. |
*Add CdCl<sub>2</sub> and FeCl<sub>2</sub> for final concentration of 100Ul = μM in each culture. | *Add CdCl<sub>2</sub> and FeCl<sub>2</sub> for final concentration of 100Ul = μM in each culture. | ||
*Incubate your labeled tubes shaking at 30°C for 2.5 hours to allow uptake. | *Incubate your labeled tubes shaking at 30°C for 2.5 hours to allow uptake. | ||
Revision as of 18:13, 23 February 2023
Contents
Introduction
Protocols
Part 1: Induce expression of Fet4_mutant
For timing reasons, the induction steps were completed prior to class. So you understand how the cell suspensions you will use for the metal uptake assay were created, please review the steps below.
- Inoculated 5 mL of SD-R media with a colony of Alpha = W303α cells transformed with Fet4_mutant.
- Incubated the culture overnight at 30 °C with shaking at 220 rpm.
- Dilute the overnight culture 1:10 in 10 mL of fresh SD-R media.
- Incubate at 30 °C for 4 hours with shaking at 220 rpm.
- To induce Fet4_mutant protein expression, pellet cells and resuspend in 10ml of SD-G media.
- Incubate overnight at at 30 °C with shaking at 220 rpm.
Part 2: Perform metal uptake experiment
- Obtain the following cultures from the front bench
- Untransformed W303α
- W303α transformed with Fet4
- W303α transformed with Fet4_mutant
- Gently tritruate with a 1ml pipette to create a homogeneous suspension for each culture
- Obtain cuvettes from the front bench to measure the OD600 of each culture prior to beginning the experiment
- Add 1ml of each cell suspension to individual cuvettes and read on the spectrophotometer
- Use 1ml SD-G for a blank.
- Record these numbers in your notebook
- Using SD-G media as a diluent, dilute each of your cultures to OD600 ~ 1.0 and a final volume of 8ml. This does not have to be exact, but the cultures should have a similar OD600 before you begin the experiment.
- Add CdCl2 and FeCl2 for final concentration of 100Ul = μM in each culture.
- Incubate your labeled tubes shaking at 30°C for 2.5 hours to allow uptake.
- During this incubation time, complete Parts 3 and 4 of the wiki.
- Following incubation, take an additional OD600 reading to account for any changes in culture density, and allow normalization of data across groups.
- Record this in your notebook.
- Transfer your cultures to 15ml conical tubes.
- Centrifuge cultures at 1000 xg for 5 minutes to pellet cells.
- During centrifugation, label 2 metal-free 15ml conical tubes for each culture, and prepare a 10ml syringe filter for each culture
- Using serological pipette, remove 6ml media supernatent without disrupting the pellet and add it to a metal-free conical tube.
- Bring your samples to the front bench and add 175ul ultra-pure nitric acid to each sample. Return to your bench with your samples.
- Using a serological pipette, carefully triturate each sample to fully mix the acid.
- Open your 10ml filter syringe and place it over the top of a fresh metal-free conical tube. Add the mixed sample to the open syringe and use the plunger to push the sample through the 0.22 μM filter.
- Close each tube with sample and place them at the front bench.
- Discard materials used to filter the samples in the black bin at the front bench.
In your laboratory notebook, complete the following:
- What volume of CdCl2 will you add to obtain a final concentration of 100 μM?
- What volume of FeCl2 will you add to obtain a final concentration of 100 μM?
- Why is it important to use metal-free tubes to store your samples for analysis?
- Why is it important to filter your samples before submitting them for analysis?
Part 3: Image Fet_mutant expression experiment
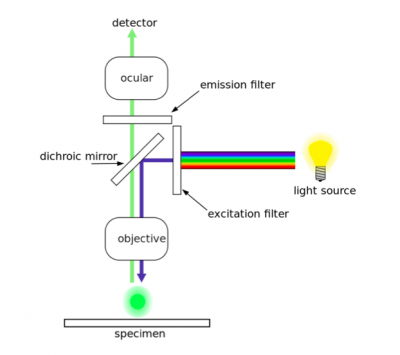
As discussed in prelab, two antibodies were used in the H2AX assay. The first antibody, or primary antibody, was anti-His tag and raised in a mouse. The secondary antibody was anti-mouse and raised in a goat, more importantly, this molecule is conjugated to a fluorescent dye tag called Alexa Fluor 568. The Alexa Fluor 568 tag is a orange/red fluorescent dye that is excited at 568 nm. To visualize the expression of your tagged mutant Fet4, we will use fluorescence microscopy.In fluorescence microscopy the specimen is illuminated with a wavelength of light specific to the excitation of the fluorescent tag used to target the feature of interest. The excitation wavelength is absorbed by the fluorescent tag, which causes it to emit light at a longer, less energetic wavelength. Typically, fluorescence microscopes used in biology are an epifluorescence type with a single light path (the objective) for excitation and emission detection, as depicted in the diagram above.
Fluorescence, or epifluorescence, microscopes are composed of a light source, an excitation filter, a dichroic mirror, and an emission filter. The filters and the dichroic mirror are specific to the spectral excitation and emission characteristics of the fluorescent tag. To visualize fluorescence, light at the excitation wavelength is focused on the sample. The light emission from the sample is focused by the objective to a detector.
Due to timing reasons, the Instructors will complete the imaging for the coverslips prepared during this laboratory session. To ensure you are familiar with the imaging procedure, each team will see a demonstration provided by the Instructor.
In your laboratory notebook, complete the following:
- What is the type of microscope used?
- What is the light source used by this microscope?
- Which objective are you using to image the yeast?
- Where do you expect to see the 568 nm signal in your images? What might it mean if you don't see the signal where it is expected? What might it mean if you see the signal where it is not expected?
Part 4: Examine Fet4 mutant expression
Reagents list
- Synthetic dropout - uracil (SD-U) media: 0.17% yeast nitrogen base without amino acid and ammonium sulfate (BD Bacto), 0.5% ammonium sulfate (Sigma), 0.192 % amino acid mix lacking uracil (Sigma), 2% glucose (BD Bacto), 0.1% adenine hemisulfate (Sigma)
- Synthetic dropout - raffinose (SD-R) media: 0.17% yeast nitrogen base without amino acid and ammonium sulfate (BD Bacto), 0.5% ammonium sulfate (Sigma), 0.192 % amino acid mix lacking uracil (Sigma), 2% raffinose (Sigma), 0.1% adenine hemisulfate (Sigma)
- Synthetic dropout - galactose (SD-G) media: 0.17% yeast nitrogen base without amino acid and ammonium sulfate (BD Bacto), 0.5% ammonium sulfate (Sigma), 0.192 % amino acid mix lacking uracil (Sigma), 2% raffinose (Sigma), 2% galactose (Sigma), 0.1% adenine hemisulfate (Sigma)
- Cadmium chloride (Sigma), stock concentration= 100mM
- Iron chloride (Sigma), stock concentration= 100mM
Next day: Confirm transporter expression and cell survival of yeast exposed to metal