Difference between revisions of "20.109(S23):M2D2"
Becky Meyer (Talk | contribs) |
(→Part 3: Use site directed mutagenesis to introduce point mutations into Fet4 transporter) |
||
| (19 intermediate revisions by one user not shown) | |||
| Line 5: | Line 5: | ||
==Introduction== | ==Introduction== | ||
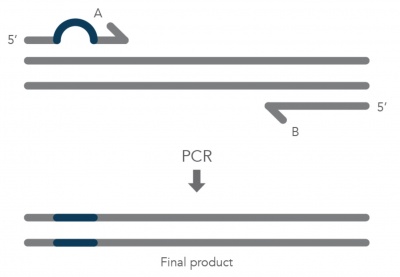
| − | + | [[Image:SDM primers IDT.jpg|right|400px|thumb|'''Schematic of primer induced point mutation in DNA replication''' Image courtesy of IDT]]In the previous laboratory session you chose your Fet4 mutagenesis strategy and generated primers to create a point mutation in the Fet4 sequence which will result in an amino acid alteration in the final protein product. Today you will perform the site-directed mutagenesis procedure necessary to generate a Fet4_mutant which you will then test. To accomplish this mutagenesis, we will use the Q5 Site-Directed Mutagenesis (SDM) kit from NEB. While there are multiple approaches to DNA mutagenesis, this kit offers the advantage of reliable generation of mutants while still being relatively cost effective. This SDM kit utilizes PCR with the specialized primers you designed to introduce and amplify the point mutation in the pFet4. | |
| − | + | Before we continue, we should review the process used to generate actual primers that are used to amplify DNA as part of this process. Current oligonucleotide, or primer, synthesis uses phosphoramidite monomers, which are simply nucleotides with protection groups added. The protection groups prevent side reactions and promote the formation of the correct DNA product. The DNA product synthesis starts with the 3'-most nucleotide and cycles through four steps: deprotection, coupling, capping, and stabilization. First, deprotection removes the protection groups. Second, during coupling the 5' to 3' linkage is generated with the incoming nucleotide. Next, a capping reaction is completed to prevent uncoupled nucleotides from forming unwanted byproducts. Lastly, stabilization is achieved through an oxidation reaction that makes the phosphate group pentavalent. For a more detailed description of this process, read [[Media:IDT chemical-synthesis-of-oligonucleotides.pdf |this article]] from IDT DNA. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
==Protocols== | ==Protocols== | ||
| − | ===Part 1: | + | ===Part 1: Research Fet4 expression plasmid=== |
| − | + | In order to study the effects of exogenous overexpression of the Fet4 transporter (or in your case, mutants of said transporter), a plasmid vector is used to express our protein of interest in yeast. The vector backbone includes several key features that enable successful expression of our mutant transporters. To understand our model system, first familiarize yourself with the important features of the expression plasmid. | |
| − | + | ||
| − | + | ||
| − | + | In this exercise, you will explore the features present in the plasmid that are necessary to express the Fet4_mutant sequence (see plasmid map below). | |
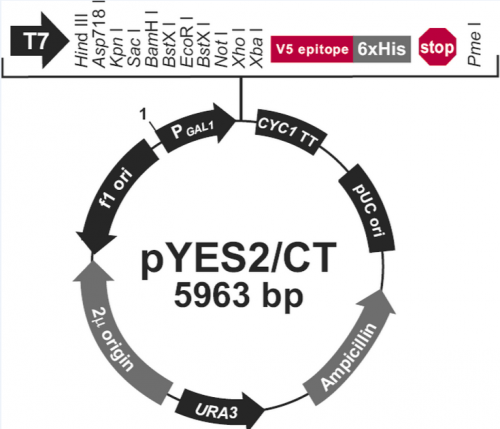
| − | + | [[Image:PYES2-CT vector map.png|right|500px|thumb|Vector map provided by Life Technologies]]<font color = #4a9152 >'''In your laboratory notebook,'''</font color> complete the following: | |
| − | + | *Describe the purpose / role for each of the following features that are present in the Fet4_mutant plasmid backbone. Please note: you many need to reference resources outside of the wiki! | |
| + | **T7 promoter | ||
| + | **Multiple cloning site | ||
| + | **V5 epitope | ||
| + | **AmpR | ||
| + | **URA3 | ||
| + | *Our expression vector is known as a "shuttle vector". What is this term, and what features of our vector enable this performance? | ||
| − | ===Part 2: | + | ===Part 2: Prepare Fet4_mutant oligos=== |
| − | + | ||
| − | + | While you were away the sequences for the mutagenesis primers you designed were submitted to Genewiz. Genewiz synthesized the DNA oligos then lyophilized (dried) it to a powder. Follow the steps below to resuspend your oligo (or 'primer'). | |
| + | #Centrifuge the tubes containing your lyophilized Fet4_mutant oligos for 1 min. | ||
| + | #Calculate the amount of water needed to give a stock concentration of 100 μM for each oligo. | ||
| + | #Resuspend each primer stock in the appropriate volume of sterile water, vortex, and centrifuge. | ||
| + | #Calculate the volume of your stock that is required to prepare a 20 μL of solution that contains your mutagenesis oligo at a concentration of 10 μM. | ||
| + | #*Try the calculation on your own first. If you get stuck, ask the teaching faculty for help. | ||
| + | #Prepare a primer mix that contains both your forward and reverse oligos at a final concentration of 10 μM in 20 μL of sterile water. | ||
| + | #*Be sure to change tips between primers! | ||
| + | #Return the rest of your Fet4_mutant oligo stocks, plus your primer specification sheet, to the front bench. | ||
| − | + | ===Part 3: Use site directed mutagenesis to introduce point mutations into Fet4 transporter=== | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | To perform site-directed mutagenesis (SDM), custom designed oligonucleotides, or primers, are used to incorporate mutations into double-stranded DNA plasmid as a specific location in the sequence. One approach is to use primers that align to the sequence in the plasmid in a back-to-back orientation. As shown in top left of the schematic below, the primers (forward primer = black arrow and reverse primer = red arrow) anneal to the plasmid such that the 5' ends of the primers anneal to the DNA in a back-to-back orientation. In Step #1 of the schematic, the forward primer is used to replicate the top strand (outside circle of the plasmid) and the reverse primer is used to replicate the bottom strand (inside circle of the plasmid). The resulting single-stranded products (extension from each primer generates a single-stranded product) are able to anneal due to sequence homology, as shown in the first quadrant of the zoom-in for Step #2. In Step #2A the 5' ends of the linear, single-stranded amplification products are phosphorylated to prepare for ligation (Step #2B). Remember that a 5' phosphate is required for 3' OH nucleophilic attack, this results in circular plasmids. | |
| − | + | Thus far in this description of SDM, one very important detail has not been mentioned. How specifically are the mutations coded in the primers incorporated into the plasmid sequence? In the top left of the schematic, the forward primer contains a hash mark that represents the desired mutation. The single-stranded product that results from extension from this primer will contain the desired mutation and therefore be incorporated into the products generated in Step #1. Lastly, in Step #2C the plasmid template that contains the unmutated parental sequence is destroyed so that only the plasmids with the desired mutation are present at the end of the procedure. | |
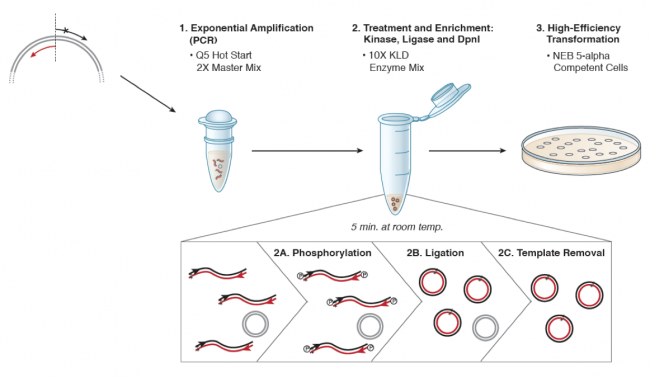
| − | [[Image:Sp16 | + | [[Image:Sp16 M1D3 SDM schematic.png|thumb|center|650px|'''Schematic of NEB Q5 Site Directed Mutagenesis procedure.''' Image modified from Q5 Site-Directed Mutagenesis Kit Manual published by NEB.]] |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | |||
| − | |||
| − | |||
| − | + | For this procedure we are using the Q5 Site Directed Mutagenesis Kit from NEB. A more technical depiction of the protocol you will use to introduce a point mutation in Fet4 is included below. Briefly, in Step 1 DNA polymerase copies the plasmid using the Fet4_mutant forward primer to insert the point mutation. Following PCR amplification the product is a linear DNA fragment. In Step 2 circular plasmids that carry the point mutation are generated when the double-stranded DNA is phosphorylated (Step 2A) and then ligated (Step 2B). Following the amplification reaction, the expression plasmid template that does not contain insert is present in the reaction product. To ensure that only the Fet4_mutant-containing expression plasmid is used in the next steps, the parental DNA is selectively digested using the DpnI enzyme (Step 2C). The underlying selective property is that DpnI only digests methylated DNA. Because DNA is methylated during replication in host cells, DNA that is synthetically made via an amplification reaction using PCR is not methylated. Lastly, in Step 3 the Fet4_mutant-containing expression plasmid is transformed into competent cells that propagate the plasmid. | |
| − | + | Each group will set up one reaction. You should work quickly but carefully, and keep your tube in a chilled container at all times. '''Please return shared reagents to the ice bucket(s) from which you took them as soon as you are done with each one.''' | |
| + | #Retrieve one PCR tube from the front laboratory bench and label the top with your team color and lab section (write small!). | ||
| + | #Add 10.25 μL of nuclease-free water. | ||
| + | #Add 1.25 μL of your primer mix (each primer should be at a concentration of 10 μM). | ||
| + | #Add 1 μL of Fet4 template plasmid DNA (concentration of 25 ng/μL). | ||
| + | #Lastly, use a filter tip to add 12.5 μL of Q5 Hot Start High-Fidelity 2X Master Mix - containing buffer, dNTPs, and polymerase - to your tube. | ||
| + | #Once all groups are ready, we will begin the thermocycler, under the following conditions: | ||
| − | + | <center> | |
| + | {| border="1" | ||
| + | ! Segment | ||
| + | ! Cycles | ||
| + | ! Temperature | ||
| + | ! Time | ||
| + | |- | ||
| + | | Initial denaturation | ||
| + | | 1 | ||
| + | | 98 °C | ||
| + | | 30 s | ||
| + | |- | ||
| + | | Amplification | ||
| + | | 25 | ||
| + | | 98 °C | ||
| + | | 10 s | ||
| + | |- | ||
| + | | | ||
| + | | | ||
| + | | 55 °C | ||
| + | | 30 s | ||
| + | |- | ||
| + | | | ||
| + | | | ||
| + | | 72 °C | ||
| + | | 2 min | ||
| + | |- | ||
| + | | Final extension | ||
| + | | 1 | ||
| + | | 72 °C | ||
| + | | 2 min | ||
| + | |- | ||
| + | | Hold | ||
| + | | 1 | ||
| + | | 4 °C | ||
| + | | indefinite | ||
| + | |} | ||
| + | </center> | ||
| − | + | After the cycling is completed, you will complete the KLD reaction (which stands for "kinase, ligase, ''Dpn''I"). | |
| − | + | #Add the following reagents: | |
| − | + | #*1 μL of your amplification product | |
| + | #*5 μL 2X KLD Reaction Buffer | ||
| + | #*1 μL KLD Enzyme Mix | ||
| + | #*3 μL nuclease-free water | ||
| + | #Incubate the reaction for 5 min at room temperature. | ||
| + | #Then, use 5 μL of the KLD reaction product to complete a transformation into an ''E. coli'' strain (NEB 5α cells of genotype ''fhuA2 Δ(argF-lacZ)U169 phoA glnV44 Φ80 Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17''). | ||
| + | #*The transformed cells will amplify the plasmid such that you are able to confirm the appropriate mutation was incorporated. | ||
| + | #Transform the cells using the following procedure: | ||
| + | #*Add 5 μL of KLD mix to 50 μL of chemically-competent NEB 5α. | ||
| + | #*Incubate on ice for 30 min. | ||
| + | #*Heat shock at 42 °C for 30 s. | ||
| + | #*Incubate on ice for 5 min. | ||
| + | #*Add 950 μL SOC and gently shake at 37 °C for 30 min. | ||
| + | #*Spread 150 μL onto LB+Amp plate and incubate overnight at 37 °C. | ||
| − | + | ==Reagents list== | |
| + | *Q5 Site Directed Mutagenesis Kit (from NEB) | ||
| + | **Q5 Hot Start High-Fidelity 2X Master Mix: propriety mix of Q5 Hot Start High-Fidelity DNA Polymerase, buffer, dNTPs, and Mg<sup>2+</sup> | ||
| + | **2X KLD Reaction Buffer | ||
| + | **10X KLD Enzyme Mix: proprietary mix of kinase, ligase, and ''DpnI'' enzymes | ||
| + | *SOC medium: 2% tryptone, 0.5% yeast extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, and 20 mM glucose | ||
| + | *LB+Amp plates | ||
| + | **Luria-Bertani (LB) broth: 1% tryptone, 0.5% yeast extract, and 1% NaCl | ||
| + | **Plates prepared by adding 1.5% agar and 100 μg/mL ampicillin (Amp) to LB | ||
| − | + | ==Navigation links== | |
| − | + | Next day: [[20.109(S23):M2D3 |Sequence clones and transform into yeast cells]] <br> | |
| − | + | Previous day: [[20.109(S23):M2D1 |Determine mutagenesis strategy]] <br> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
Latest revision as of 14:25, 17 March 2023
Contents
Introduction
In the previous laboratory session you chose your Fet4 mutagenesis strategy and generated primers to create a point mutation in the Fet4 sequence which will result in an amino acid alteration in the final protein product. Today you will perform the site-directed mutagenesis procedure necessary to generate a Fet4_mutant which you will then test. To accomplish this mutagenesis, we will use the Q5 Site-Directed Mutagenesis (SDM) kit from NEB. While there are multiple approaches to DNA mutagenesis, this kit offers the advantage of reliable generation of mutants while still being relatively cost effective. This SDM kit utilizes PCR with the specialized primers you designed to introduce and amplify the point mutation in the pFet4.Before we continue, we should review the process used to generate actual primers that are used to amplify DNA as part of this process. Current oligonucleotide, or primer, synthesis uses phosphoramidite monomers, which are simply nucleotides with protection groups added. The protection groups prevent side reactions and promote the formation of the correct DNA product. The DNA product synthesis starts with the 3'-most nucleotide and cycles through four steps: deprotection, coupling, capping, and stabilization. First, deprotection removes the protection groups. Second, during coupling the 5' to 3' linkage is generated with the incoming nucleotide. Next, a capping reaction is completed to prevent uncoupled nucleotides from forming unwanted byproducts. Lastly, stabilization is achieved through an oxidation reaction that makes the phosphate group pentavalent. For a more detailed description of this process, read this article from IDT DNA.
Protocols
Part 1: Research Fet4 expression plasmid
In order to study the effects of exogenous overexpression of the Fet4 transporter (or in your case, mutants of said transporter), a plasmid vector is used to express our protein of interest in yeast. The vector backbone includes several key features that enable successful expression of our mutant transporters. To understand our model system, first familiarize yourself with the important features of the expression plasmid.
In this exercise, you will explore the features present in the plasmid that are necessary to express the Fet4_mutant sequence (see plasmid map below).
In your laboratory notebook, complete the following:- Describe the purpose / role for each of the following features that are present in the Fet4_mutant plasmid backbone. Please note: you many need to reference resources outside of the wiki!
- T7 promoter
- Multiple cloning site
- V5 epitope
- AmpR
- URA3
- Our expression vector is known as a "shuttle vector". What is this term, and what features of our vector enable this performance?
Part 2: Prepare Fet4_mutant oligos
While you were away the sequences for the mutagenesis primers you designed were submitted to Genewiz. Genewiz synthesized the DNA oligos then lyophilized (dried) it to a powder. Follow the steps below to resuspend your oligo (or 'primer').
- Centrifuge the tubes containing your lyophilized Fet4_mutant oligos for 1 min.
- Calculate the amount of water needed to give a stock concentration of 100 μM for each oligo.
- Resuspend each primer stock in the appropriate volume of sterile water, vortex, and centrifuge.
- Calculate the volume of your stock that is required to prepare a 20 μL of solution that contains your mutagenesis oligo at a concentration of 10 μM.
- Try the calculation on your own first. If you get stuck, ask the teaching faculty for help.
- Prepare a primer mix that contains both your forward and reverse oligos at a final concentration of 10 μM in 20 μL of sterile water.
- Be sure to change tips between primers!
- Return the rest of your Fet4_mutant oligo stocks, plus your primer specification sheet, to the front bench.
Part 3: Use site directed mutagenesis to introduce point mutations into Fet4 transporter
To perform site-directed mutagenesis (SDM), custom designed oligonucleotides, or primers, are used to incorporate mutations into double-stranded DNA plasmid as a specific location in the sequence. One approach is to use primers that align to the sequence in the plasmid in a back-to-back orientation. As shown in top left of the schematic below, the primers (forward primer = black arrow and reverse primer = red arrow) anneal to the plasmid such that the 5' ends of the primers anneal to the DNA in a back-to-back orientation. In Step #1 of the schematic, the forward primer is used to replicate the top strand (outside circle of the plasmid) and the reverse primer is used to replicate the bottom strand (inside circle of the plasmid). The resulting single-stranded products (extension from each primer generates a single-stranded product) are able to anneal due to sequence homology, as shown in the first quadrant of the zoom-in for Step #2. In Step #2A the 5' ends of the linear, single-stranded amplification products are phosphorylated to prepare for ligation (Step #2B). Remember that a 5' phosphate is required for 3' OH nucleophilic attack, this results in circular plasmids.
Thus far in this description of SDM, one very important detail has not been mentioned. How specifically are the mutations coded in the primers incorporated into the plasmid sequence? In the top left of the schematic, the forward primer contains a hash mark that represents the desired mutation. The single-stranded product that results from extension from this primer will contain the desired mutation and therefore be incorporated into the products generated in Step #1. Lastly, in Step #2C the plasmid template that contains the unmutated parental sequence is destroyed so that only the plasmids with the desired mutation are present at the end of the procedure.
For this procedure we are using the Q5 Site Directed Mutagenesis Kit from NEB. A more technical depiction of the protocol you will use to introduce a point mutation in Fet4 is included below. Briefly, in Step 1 DNA polymerase copies the plasmid using the Fet4_mutant forward primer to insert the point mutation. Following PCR amplification the product is a linear DNA fragment. In Step 2 circular plasmids that carry the point mutation are generated when the double-stranded DNA is phosphorylated (Step 2A) and then ligated (Step 2B). Following the amplification reaction, the expression plasmid template that does not contain insert is present in the reaction product. To ensure that only the Fet4_mutant-containing expression plasmid is used in the next steps, the parental DNA is selectively digested using the DpnI enzyme (Step 2C). The underlying selective property is that DpnI only digests methylated DNA. Because DNA is methylated during replication in host cells, DNA that is synthetically made via an amplification reaction using PCR is not methylated. Lastly, in Step 3 the Fet4_mutant-containing expression plasmid is transformed into competent cells that propagate the plasmid.
Each group will set up one reaction. You should work quickly but carefully, and keep your tube in a chilled container at all times. Please return shared reagents to the ice bucket(s) from which you took them as soon as you are done with each one.
- Retrieve one PCR tube from the front laboratory bench and label the top with your team color and lab section (write small!).
- Add 10.25 μL of nuclease-free water.
- Add 1.25 μL of your primer mix (each primer should be at a concentration of 10 μM).
- Add 1 μL of Fet4 template plasmid DNA (concentration of 25 ng/μL).
- Lastly, use a filter tip to add 12.5 μL of Q5 Hot Start High-Fidelity 2X Master Mix - containing buffer, dNTPs, and polymerase - to your tube.
- Once all groups are ready, we will begin the thermocycler, under the following conditions:
| Segment | Cycles | Temperature | Time |
|---|---|---|---|
| Initial denaturation | 1 | 98 °C | 30 s |
| Amplification | 25 | 98 °C | 10 s |
| 55 °C | 30 s | ||
| 72 °C | 2 min | ||
| Final extension | 1 | 72 °C | 2 min |
| Hold | 1 | 4 °C | indefinite |
After the cycling is completed, you will complete the KLD reaction (which stands for "kinase, ligase, DpnI").
- Add the following reagents:
- 1 μL of your amplification product
- 5 μL 2X KLD Reaction Buffer
- 1 μL KLD Enzyme Mix
- 3 μL nuclease-free water
- Incubate the reaction for 5 min at room temperature.
- Then, use 5 μL of the KLD reaction product to complete a transformation into an E. coli strain (NEB 5α cells of genotype fhuA2 Δ(argF-lacZ)U169 phoA glnV44 Φ80 Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17).
- The transformed cells will amplify the plasmid such that you are able to confirm the appropriate mutation was incorporated.
- Transform the cells using the following procedure:
- Add 5 μL of KLD mix to 50 μL of chemically-competent NEB 5α.
- Incubate on ice for 30 min.
- Heat shock at 42 °C for 30 s.
- Incubate on ice for 5 min.
- Add 950 μL SOC and gently shake at 37 °C for 30 min.
- Spread 150 μL onto LB+Amp plate and incubate overnight at 37 °C.
Reagents list
- Q5 Site Directed Mutagenesis Kit (from NEB)
- Q5 Hot Start High-Fidelity 2X Master Mix: propriety mix of Q5 Hot Start High-Fidelity DNA Polymerase, buffer, dNTPs, and Mg2+
- 2X KLD Reaction Buffer
- 10X KLD Enzyme Mix: proprietary mix of kinase, ligase, and DpnI enzymes
- SOC medium: 2% tryptone, 0.5% yeast extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, and 20 mM glucose
- LB+Amp plates
- Luria-Bertani (LB) broth: 1% tryptone, 0.5% yeast extract, and 1% NaCl
- Plates prepared by adding 1.5% agar and 100 μg/mL ampicillin (Amp) to LB
Next day: Sequence clones and transform into yeast cells