20.109(S21):M3D1
Contents
Introduction
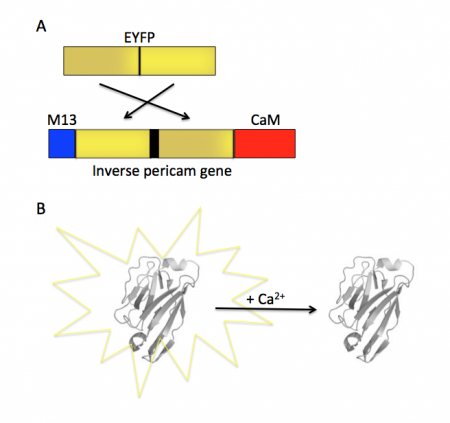
Today you will familiarize yourself with the recombinant protein IPC and its constituent parts. The fluorescent component of IPC is an enhanced yellow fluorescent protein (abbreviated EYFP), one of the many derivatives of green fluorescent protein (GFP). GFP is naturally produced by jellyfish and was cloned into other organisms in the early 1990’s. It has since been exploited as a genetically encodable reporter and mutagenized to vary its excitation and emission spectra. The other key component of inverse pericam is the protein calmodulin (CaM), a natural calcium sensor that is present in all eukaryotes Calmodulin has many ligands that it binds only in the presence of calcium ion, including the peptide fragment M13. This conditional specificity for M13 binding is enabled by the change in confirmation of CaM when bound to calcium.
Within inverse pericam, M13 and CaM are located at opposite ends, surrounding a permuted (i.e., rearranged) version of EYFP. In the absence of calcium, this EYFP exhibits strong fluorescence. However, when enough calcium is added to a solution of inverse pericam, CaM and M13 interact, disrupting the conformation and, as a result, the fluorescence of EYFP. The transition from bright to dim fluorescence occurs over a particular concentration range of calcium. The calcium concentration at which binding to CaM occurs (and fluorescence decreases) is referred to as the Kd and determined by the affinity of CaM to calcium. In addition, the interaction between CaM and calcium is impacted by cooperativity. CaM has four calcium binding sites. In cooperativity, the affinity of CaM for calcium is altered by how many calcium ions are already bound to the protein. The mutations you will examine today were designed in an effort to modify the calcium sensor portion of IPC in a manner that is likely to change the affinity and / or cooperativity for calcium ions.
To examine the modification that were made to IPC, we will use several protein analysis tools. Proteins are modular materials that may be described and examined at multiple levels of a structural hierarchy (from primary to quaternary in the classical paradigm). Primary structure refers to a protein’s amino acid sequence, which might reveal a cluster of charged residues or a pattern of alternating polar and nonpolar residues. One cannot predict off-hand the conformation of a protein merely from its linear sequence; however, due to rotational flexibility of bonds and non-covalent interactions between non-adjacent amino acids (as well as covalent disulfide bonds) some structural characteristics can be inferred. Because many proteins have structural motifs in common (e.g., alpha helices and beta sheets at the secondary level, or leucine-rich repeats at the tertiary level), which ultimately arise from the amino acid sequences, databases can be useful for making predictions about proteins with known amino acid sequences but unknown structures.
Protocols
Part 1: Review IPC reference article
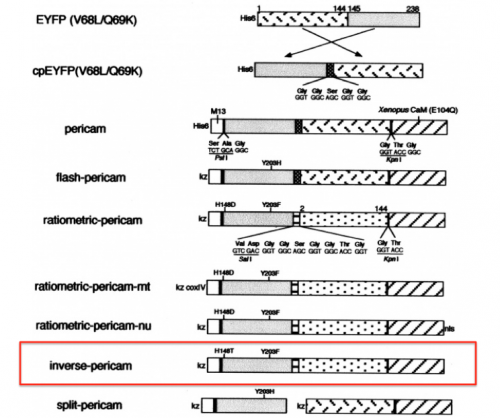
Previous 109ers generated the data you will analyze in this module by changing specific amino acids in the IPC protein sequence. The goal of this directed mutagenesis approach was to alter the interaction between IPC and calcium such that the affinity / cooperativity was improved. Before we examine the effect of these mutations on the activity of IPC, we will first review how the original IPC sensor was constructed.
With your partner, review the information regarding the development of calcium sensor variants in the paper by Nagai et. al. (attached here).
Part 2: Identify IPC sequence features
- Open your SnapGene file of the IPC sequence and label the features listed below.
- M13 peptide: 1-78 bp
- EYFP (C-terminus portion): 91-372 bp
- EYFP (N-terminus portion): 400-831 bp
- CaM: 838-1281 bp
- Linker sequences: 82-90, 373-399, and 832-837 bp
- Refer to the figure shown above, which depicts the IPC construct in schematic form, to assist in your understanding of how the different components of IPC are connected.
- To understand how the mutations in the IPC variants might effect calcium binding, it is helpful to identify features within the IPC sequence that are important to the functionality of the protein. For this, it is best to translated protein sequence rather than the gene sequence.
- The amino acids are shown in the SnapGene sequence window below the DNA bases for the coding regions.
- To assist in the identification of key features in the IPC sequence, review the information provided in the paper by Zhang et. al. (linked here). In particular, carefully read the following: Abstract, Introduction, and the "Linker and loop flexibility" section in the Results.
- In your IPC SnapGene file, label the amino acid residues that comprise the calcium-binding loops in the CaM region of IPC.
- If you get stuck, use the fact that the CaM within inverse pericam is an E103Q mutant, that is, the 103rd residue of calmodulin is Q, to keep yourself oriented.
- Consider other regions of CaM that might be important for calcium binding and label in your IPC SnapGene file.
- Perhaps the "Loss of hydrophobic cavities" section in the Results will provide interesting potential targets.
- In your laboratory notebook, complete the following:
- Upload the labeled IPC gene sequence and the labeled IPC protein sequence.
- Examine the four calcium binding loops. Do these loops share any common features? Do any of these loops contain unique features?
- What additional regions did you mark as interesting? Why?
- Suggest a mutation that you think might impact the activity of IPC (be specific, what amino acid will replace what amino acid?). Do you hypothesize that this mutation will increase or decrease the affinity of calcium binding? Why? Do you hypothesize that this mutation will increase or decrease the cooperativity of calcium binding? Why?
Part 3: Examine IPC structural elements
In the previous section you reviewed primary scientific literature to locate important features in the IPC sequence. Now you will examine 3D representations of CaM to visualize those features more closely.
- You will examine the structure of CaM using the Protein Data Bank (PDB) (linked here). In this online database, the structures are organized according to PDB identification codes.
- To begin, you will look at the calcium-bound form of CaM.
- Enter "1CLL" into the search box at the top right corner of the PDB homepage.
- The program will open in FirstView mode for the structure you’ve chosen (ensure that popup blockers are off if the structure fails to load). On the right is the image panel, which shows your protein along with associated ligands (in this case, calcium). Try clicking and dragging on the rotating image to see what happens.
- Now look at the control panel on the upper left: here you can modify the image. Try adding and removing water molecules and ligands see where they interact with the protein.
- As you explore the features of the control panel and image panel, be sure to observe the message frame window on the lower left for any relevant information that may pop up. If you click on an atom in the image panel, its atomic identity will be displayed in the message frame, along with its encompassing amino acid residue and position.
- From the control panel, click on the PDB icon, which leads to detailed information about the publication upon which the model image is based.
- To find further options for modifying how you view the image, or search for particular atoms, click on More Views in the control panel, or on Jmol at the bottom right of the image panel. For example, you can highlight specific amino acids, or change from a backbone trace to a space-filling model. Explore these features. For example, you might use color to highlight all the acidic amino acids in calmodulin.
- Be sure to note any useful information in your notebook as you go. You might ask:
- What method was used to elucidate the structure of this protein?
- How good is the image resolution?
- Which species did this protein come from?
- When did the authors publish their results?
- What are the major components of the molecule’s secondary structure?
- What do the calcium binding loops (or other areas of interest you found) look like?
- Once you are satisfied with your understanding of calcium-bound calmodulin, look at an apo calmodulin structure (or two) for comparison. You might find the structure directly by using PDB, or by using the NCBI Structure database.
- Write a few sentences in your lab notebook describing the differences between the calcium-bound and apo forms of calmodulin.
Next day: Identify IPC mutations