20.109(S21):M3D1
Primer design
To amplify a specific sequence of DNA, you first need to design primers -- one primer that anneals at the start of the sequence of interest (the 5' end) and a second primer that anneals at the end of the sequence of interest (the 3' end). The primer that anneals at the start of the sequence is referred to as the 'forward' primer. The forward primer anneals to the non-coding DNA strand and reads toward, or into, the gene of interest. The 'reverse' primer anneals to the coding DNA strand at the end of the sequence and reads back into the sequence. Primers can also be useful in adding sequence to sequences upon amplification via the polymerase chain reaction.
Polymerase chain reaction (PCR)
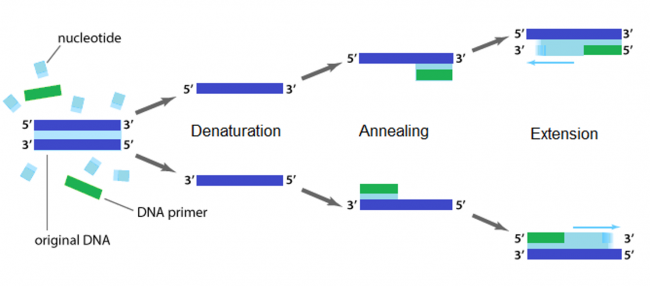
The applications of PCR are widespread, from forensics to molecular biology to evolution, but the goal of any PCR is the same: to generate many copies of DNA from a single or a few specific sequence(s) (called the “template”). In addition to the template, PCR requires only three components: primers to bind sequence flanking the target, dNTPs to polymerize, and a heat-stable polymerase to catalyze the synthesis reaction over and over and over. DNA polymerases require short initiating pieces of DNA called primers to copy DNA. In PCR amplification, forward and reverse primers that target the non-coding and coding strands of DNA, respectively, are separated by a distance equal to the length of the DNA to be copied. To amplify DNA, the original DNA segment, or template DNA, is denatured using heat. This separates the strands and allows the primers to anneal to the template. Then polymerase extends from the primer to copy the template DNA. How many cycles of PCR are required to achieve the desired double-stranded amplification product?
Several features are important to consider when designing primers for PCR. Primers that are too short may lack requisite specificity for the desired sequence, and thus amplify an unrelated sequence. The longer a primer is, the more favorable are its energetics for annealing to the template DNA, due to increased hydrogen bonding. On the other hand, longer primers are more likely to form secondary structures such as hairpins, leading to inefficient template priming. Two other important features are G/C content and placement. Having a G or C base at the end of each primer increases priming efficiency, due to the greater energy of a GC pair compared to an AT pair. The latter decrease the stability of the primer-template complex. Overall G/C content should ideally be 50 +/- 10%, because long stretches of G/C or A/T bases are both difficult to copy. The G/C content also affects the melting temperature. PCR is a three-step process (denature, anneal, extend) and these steps are repeated 20 or more times. After 30 cycles of PCR, there could be as many as a billion copies of the original template sequence.