Difference between revisions of "20.109(S17):Prepare Western blot and induce DNA damage for quantitative PCR assay (Day2)"
Noreen Lyell (Talk | contribs) (→Part 1b: Transfer proteins onto nitrocellulose membrane) |
Noreen Lyell (Talk | contribs) (→Navigation links) |
||
| Line 175: | Line 175: | ||
==Navigation links== | ==Navigation links== | ||
| − | Next day: [[20.109(S17): | + | Next day: [[20.109(S17):Purify RNA for quantitative PCR assay and treat cells for survival assay (Day3)| Purify RNA for quantitative PCR assay and treat cells for survival assay]] |
Previous day: [[20.109(S17):Confirm cell lines and practice tissue culture techniques (Day1)| Confirm cell lines and practice tissue culture techniques]] | Previous day: [[20.109(S17):Confirm cell lines and practice tissue culture techniques (Day1)| Confirm cell lines and practice tissue culture techniques]] | ||
Revision as of 03:12, 7 March 2017
Contents
Introduction
In the previous laboratory session you isolated the protein fraction from cell lysates. Today you will separate the proteins in the lysates using a polyacrylamide gel and transfer the proteins onto a nitrocellulose membrane. This step enables you to 'probe' the protein fractions isolated from cells for your protein of interest, which in our experiment is BRCA2. To probe the membrane, it was incubated with two primary antibodies overnight by the teaching faculty - α-BRCA2 and α-α-tubulin. The α-BRCA2 antibody was raised in a XXXXX and the α-α-tubulin antibody was raised in a rabbit. Primary antibodies bind directly to the protein of interest. Today you will add a secondary antibodies. Each secondary antibody will bind to the appropriate primary antibody and provide a means for visualization - in our experiment, the secondary antibody is responsible for a fluorescent signal...think back to the anti-His antibody used in module 1!
The ability to bind specific proteins using antibodies, or immunoglobulins, is critical in Western blot analysis. Antibodies are typically 'raised' in mammalian hosts. Most commonly mice, rabbits, and goats are used, but antibodies can also be raised in sheep, chickens, rats, and even humans. The protein used to raise an antibody is called the antigen and the portion of the antigen that is recognized by an antibody is called the epitope. Some antibodies are monoclonal, or more appropriately “monospecific,” and recognize one epitope, while other antibodies, called polyclonal antibodies, are in fact antibody pools that recognize multiple epitopes. Antibodies can be raised not only to detect specific amino acid sequences, but also post-translational modifications and/or secondary structure. Therefore, antibodies can be used to distinguish between modified (for example, phosphorylated or glycoslyated proteins) and unmodified protein.
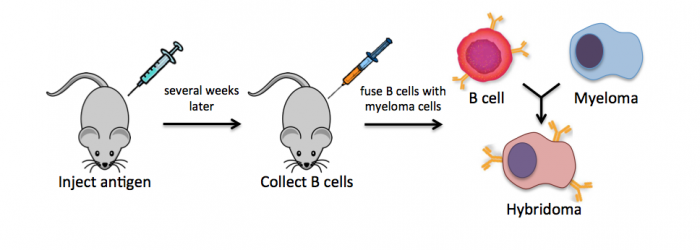
Monoclonal antibodies overcome many limitations of polyclonal pools in that they are specific to a particular epitope and can be produced in unlimited quantities. However, more time is required to establish these antibody-producing cells, called hybridomas, and it is a more expensive endeavor. In this process, normal antibody-producing B cells are fused with immortalized B cells, derived from myelomas, by chemical treatment with a limited efficiency. To select only heterogeneously fused cells, the cultures are maintained in medium in which myeloma cells alone cannot survive (often HAT medium). Normal B cells will naturally die over time with no intervention, so ultimately only the fused cells, called hybridomas, remain. A fused cell with two nuclei can be resolved into a stable cell line after mitosis.
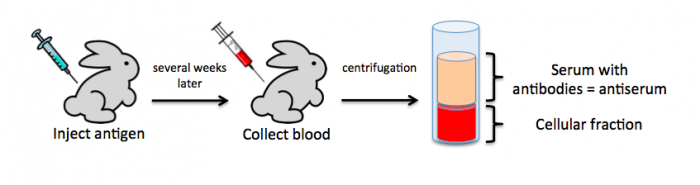
To raise polyclonal antibodies, the antigen of interest is first purified and then injected into an animal. To elicit and enhance the animal’s immunogenic response, the antigen is often injected multiple times over several weeks in the presence of an immune-boosting compound called adjuvant. After some time, usually 4 to 8 weeks, samples of the animal’s blood are collected and the cellular fraction is removed by centrifugation. What is left, called the serum, can then be tested in the lab for the presence of specific antibodies. Even the very best antisera have no more than 10% of their antibodies directed against a particular antigen. The quality of any antiserum is judged by the purity (that it has few other antibodies), the specificity (that it recognizes the antigen and not other spurious proteins) and the concentration (sometimes called titer). Animals with strong responses to an antigen can be boosted with the antigen and then bled many times, so large volumes of antisera can be produced. However animals have limited life-spans and even the largest volumes of antiserum will eventually run out, requiring a new animal. The purity, specificity and titer of the new antiserum will likely differ from those of the first batch. High titer antisera against bacterial and viral proteins can be particularly precious since these antibodies are difficult to raise; most animals have seen these immunogens before and therefore don’t mount a major immune response when immunized. Antibodies against toxic proteins are also challenging to produce if they make the animals sick.
For Western blot analysis, a high quality antibody can have a relatively low affinity for its target protein. This is because the target is localized and concentrated on a blot, allowing the antibody to bind using both antibody “arms” thereby strengthening the association. Even an antibody that is loosely bound to the blot under these circumstances may dissociate then re-associate quickly since the local concentration of the target protein is high. The lower limit for protein detection is approximately 1 ng/lane, a value that varies with the size of the protein to be detected and the Western blotting apparatus that is used. For most polyacrylamide gels, the protein capacity for each lane is 100 to 200 μg (that would be 20 μL of a 5-10 μg/μL protein preparation). Thus, 1 ng represents a protein that is approximately 0.0005-0.001% of the total cellular protein (1 ng out of 100,000-200,000 ng). Proteins that make up a more significant fraction of the total protein population will be easier to detect.
Traditionally, only one species of antibody could be used on a Western blot because the detection relied on the emission of light that was collected by x-ray film. In the traditional systems the output looks like black bands on a blue or clear background. However, more recent conjugate chemistry has allowed secondary antibodies to be coupled to fluorescent tags. Today we will use infrared (IR) secondary antibodies to detect our α-BRCA2 and α-tubulin antibodies and then scan the Western blots using a specially constructed microscope located in the Lauffenburger lab to determine the level of BRCA2 in our cell lines.
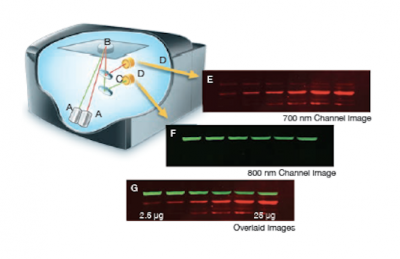
The Licor Odyssey scanner consists of an inverted microscope with two lasers that excite dyes which emit light in the IR range. As depicted in the image on the right, an excitation point is created when beams from the 700 nm and 800 nm lasers (A) are focused on the scanning surface. The microscope objective (B) is focused on the excitation point and collects light from the fluorescing IR dyes. This light is passed through a dichroic mirror (C) that separates the light into two distinct signals that travel through two independent optical paths that are focused on separate silicon photodiodes (D) and detected. In the image, the first channel (E) and second channel (F) are shown separately and merged (G).We will detect our IR-dye conjugated secondary antibodies at wavelengths of 700 and 800 nm. The 700 nm channel will appear red and the 800 nm channel will appear green. Infrared secondary antibodies provide a more flexible detection platform than the traditional Western blot detection methods that rely on colorimetric or chemiluminescent substrates. Unlike the colorimetric or chemiluminescent detection methods, IR dyes do not require a chemical reaction to occur in order for signal to be detected. This means that the output signal increases with time as the colorimetric or chemiluminescent substrate reaction proceeds -- making timing an important variable in traditional Western blot development. We remove that variable from the equation and control when we want to visualize our Western blot simply by controlling the excitation of the dye.
Protocols
Part 1: Prepare Western blot
Last time, you prepared protein extracts from DLD-1 and BRCA2- cells and they were stored at -20 °C.
Part 1a: Separate cell lysate proteins using SDS-PAGE
- Retrieve your cell lysate samples from the front laboratory bench.
- Centrifuge your sample for 1 min at max speed.
- Load your samples according to the scheme below.
- In your notebook, document the start and stop time of electrophoresis.
- The teaching faculty will begin electrophoresis after both groups load their samples.
- The proteins will be separated using 200 V for 35 minutes.
| Lane | Sample | Volume to load |
|---|---|---|
| 1 | EMPTY | N/A |
| 2 | Team 1, DLD-1 | 20 μL |
| 3 | Team 1, BRCA2- | 20 μL |
| 4 | HiMark ladder | 10 μL |
| 5 | EMPTY | N/A |
| 6 | HiMark ladder | 10 μL |
| 7 | Team 2, DLD-1 | 20 μL |
| 8 | Team 2, BRCA2- | 20 μL |
| 9 | HiMark ladder | 10 μL |
| 10 | EMPTY | N/A |
Part 1b: Transfer proteins onto nitrocellulose membrane
After the electrophoresis procedure, the teaching faculty will assist you in assembling the transfer cassette according the below protocol. If we run short on time, the teaching faculty will complete this part for you.
- Wearing gloves, carefully disassemble the electrophoresis chamber.
- Assemble the transfer cassette in the following order:
- Place the black side of the transfer cassette in a tupperware container with transfer buffer. The transfer cassette is color-coded so the black side should face the cathode (black electrode) and the clear side should face the anode (red electrode).
- Place a ScotchBrite pad pre-soaked in transfer buffer on the black side of the cassette.
- Place 1 piece of filter paper on top of the ScotchBrite pad.
- Place your gel on top of the filter paper.
- Lightly press out any air bubbles that form between the filter paper and your gel.
- Place a piece of nitrocellulose membrane on top of the gel.
- The nitrocellulose membrane is white and should be kept between the blue protective paper sheets until use. Wear gloves when handling the membrane to avoid transferring proteins from your fingers to the membrane.
- Again, lightly press out any air bubbles that form between the gel and the nitrocellulose membrane.
- Place another piece of filter paper on top of the nitrocellulose.
- Place a second ScotchBrite pad pre-soaked in transfer buffer on top of the filter paper.
- Close the cassette, then push the clasp down and slide it along the top to hold it together.
- Place the transfer cassette into the blotting tank so that the clear side faces the red electrode and the black side faces the black electrode.
- Two blots can be run in each tank. When both are in place, insert a magnetic stir bar and an ice compartment into the tank and fill the tank with transfer buffer.
- Connect the power supply and transfer at 100 V for 60 min.
- After the transfer is complete, turn off the current, disconnect the blotting tank from the power supply, and remove the transfer cassettes.
- Disassemble the transfer cassette to retrieve the nitrocellulose membrane and confirm that the pre-stained standard markers transferred from the gel to the membrane.
- Cut the membrane such that each team has a membrane with only their samples, and then transfer each membrane to a separate plastic tupperware.
- Add enough Odyssey blocking buffer to the plastic dish to just cover the membrane.
- Your membranes will be stored overnight in blocking buffer at 4°C.
The night before your next laboratory session, the teaching faculty will pour off the blocking buffer and incubate your membrane in the primary antibody solution until you return.
Part 2: Induce DNA damage
To measure gene expression of DNA cell cycle checkpoint factors, you will perform quantitative PCR (qPCR). In qPCR, the amount of a specific transcript can be measured by probing the RNA sequence with primers. We will discuss the details more on M2D9.
In this exercise, you will induce DNA damage in the cells you seeded in the previous laboratory session. Next class you will purify RNA from your cells for quantitative PCR analysis.
- Prepare your working space within the tissue culture hood.
- Calculate the volume of etoposide stock needed to induce DNA damage.
- Obtain an aliquot of pre-warmed media from the 37 °C water bath (25 mL).
- Determine the volume of etoposide stock (100 mM) you need to add to the media for a final concentration of 100 μM.
- Retrieve your four T75 flasks (two DLD-1 cultures and two BRCA2- cultures) from the 37 °C incubator and visually inspect your cells with a microscope.
- Record your observations concerning media color, confluency, etc. in your laboratory notebook.
- Move your flasks into the tissue culture hood.
- Aspirate the spent media from one DLD-1 and one BRCA2- flask.
- Be careful not to cross-contaminant between flasks with different cell lines!
- Add 5 mL of PBS to each flask and rock the plate gently to wash the cells.
- Aspirate the PBS from each flask.
- Again, be careful not to cross-contaminate.
- Add 10 mL of the media containing etoposide that you prepared in Step #2.
- Carefully put your flasks in the 37 °C incubator for 60 min.
- Aspirate the spent media from the remaining (non-etoposide treated) DLD-1 and BRCA2- flasks and replace with 10 mL of fresh media, then return to the 37 °C incubator.
- Be sure to label these flasks as 'untreated control', then return the flasks to the 37 °C incubator.
- Following the 60 min DNA damage induction, aspirate the media containing etoposide from each flask.
- Be mindful of cross-contamination.
- Add 10 mL of fresh media to each flask.
- Label these flasks '+etop' to denote that the cells were treated with etoposide, then return the flasks to the 37 °C incubator.
- Clean your working space within the tissue culture hood.
Part 3: Discuss research paper
We will end today with a discussion of the Dietlein et al. research article. In their research, the authors completed a screen to examine 1319 cancer-associated genes from 67 cell lines to identify cancer-cell specific mutations that are associated with DNA-PKcs dependence or addiction. Refer to the M2 overview for more information on addiction in cancer cells.
Our paper discussion will assist you in writing a cohesive story that clearly reports the data and provides strong support for the conclusions made about the data. During the paper discussion, everyone is expected to participate - either by volunteering or by being called upon!
Introduction
Remember the key components of an introduction:
- What is the big picture?
- Is the importance of this research clear?
- Are you provided with the information you need to understand the research?
- Do the authors include a preview of the key results?
Results
Carefully examine the figures. First, read the captions and use the information to 'interpret' the data presented within the image. Second, read the text within the results section that describes the figure.
- Do you agree with the conclusion(s) reached by the authors?
- What controls are included and are they appropriate for the experiment performed?
- Are you convinced that the data are accurate and/or representative?
Discussion
Consider the following components of a discussion:
- Are the results summarized?
- Did the authors 'tie' the data together into a cohesive and well-interpreted story?
- Do the authors overreach when interpreting the data?
- Are the data linked back to the big picture from the introduction?
Reagents
Next day: Purify RNA for quantitative PCR assay and treat cells for survival assay
Previous day: Confirm cell lines and practice tissue culture techniques