Difference between revisions of "20.109(F16):Module 1"
Noreen Lyell (Talk | contribs) (→Notes for teaching faculty) |
Noreen Lyell (Talk | contribs) (→References) |
||
| Line 36: | Line 36: | ||
==References== | ==References== | ||
| − | |||
| − | |||
| − | |||
| − | |||
==Notes for teaching faculty== | ==Notes for teaching faculty== | ||
[[20.109(F16): Prep notes for module 1| Prep notes, M1(F16)]] | [[20.109(F16): Prep notes for module 1| Prep notes, M1(F16)]] | ||
[[20.109(F16): Prep notes for orientation| F16 notes for orientation day]] | [[20.109(F16): Prep notes for orientation| F16 notes for orientation day]] | ||
Revision as of 20:43, 28 June 2016
Contents
Module 1
Lecturer: Bevin Engelward
Instructors: Noreen Lyell, Leslie McClain and Maxine Jonas
TA:
Lab manager: Hsinhwa Lee
Overview
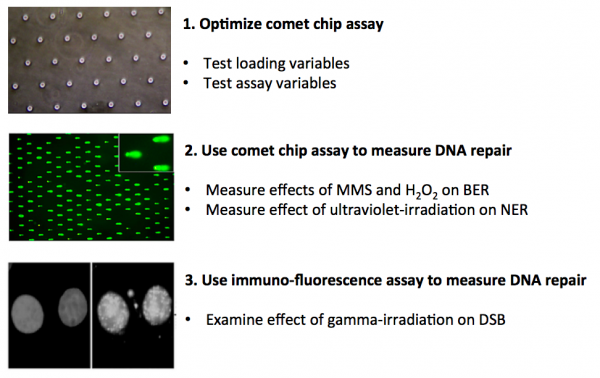
In this module you will measure DNA repair using two assays: the comet chip and immuno-fluorescence. Your first task is to critically think through the development of the comet chip assay and determine which conditions provide the best results. To this end, you will consider variables that effect cell loading into the microwells of the comet chip and variables that effect the readout of the results. The data you collect will be used to propose a high-throughput comet chip assay for commercial use.
Next, you will use the comet chip assay to assess the effect of chemicals and ultraviolet-irradiation on DNA repair. Specifically, you will study the base excision repair (BER) pathway and the nucleotide excision repair (NER) pathway. Last, you will examine the effect of gamma-irradiation on double strand breaks using an immuno-fluorescence approach.
Lab links: day by day
M1D1: DNA engineering using PCR
M1D2: Clean and cut DNA
M1D3: Agarose gel electrophoresis
M1D4: Ligation & transformation
M1D5: Examine candidate clones and tissue culture
M1D6: Lipofection
M1D7: Data analysis
Assignments
Business plan presetation
DNA repair summary