Difference between revisions of "Understanding the lock-in amplifier"
(→MATLAB Simulation) |
|||
| Line 32: | Line 32: | ||
==MATLAB Simulation== | ==MATLAB Simulation== | ||
| − | + | The following Matlab code was used to generate the figures above. | |
<pre> | <pre> | ||
| − | |||
close all | close all | ||
clear all | clear all | ||
| Line 86: | Line 85: | ||
brownNoise = step(coloredNoiseGenerator); | brownNoise = step(coloredNoiseGenerator); | ||
brownNoiseNorm = (brownNoise-min(brownNoise))/(max(brownNoise)-min(brownNoise))-0.4; | brownNoiseNorm = (brownNoise-min(brownNoise))/(max(brownNoise)-min(brownNoise))-0.4; | ||
| − | |||
% First figure demonstrates that because signal and noise occupy the same | % First figure demonstrates that because signal and noise occupy the same | ||
| Line 157: | Line 155: | ||
,'FontSize', 14) | ,'FontSize', 14) | ||
axis([-0.3 .3 1E-1 inf]) | axis([-0.3 .3 1E-1 inf]) | ||
| − | |||
% The third figure demonstrates what happens when we modulate the the | % The third figure demonstrates what happens when we modulate the the | ||
| Line 194: | Line 191: | ||
set(gca,'xtick',[-0.2,-0.1, 0, 0.1, 0.2],'xticklabel',{'-2w','-w','0','w','2w'},'ytick',[]... | set(gca,'xtick',[-0.2,-0.1, 0, 0.1, 0.2],'xticklabel',{'-2w','-w','0','w','2w'},'ytick',[]... | ||
,'FontSize', 14) | ,'FontSize', 14) | ||
| − | |||
| − | |||
% The fourth figure demonstrates what happens when you multiply the | % The fourth figure demonstrates what happens when you multiply the | ||
| Line 228: | Line 223: | ||
set(gca,'xtick',[-0.2,-0.1, 0, 0.1, 0.2],'xticklabel',{'-2w','-w','0','w','2w'},'ytick',[]... | set(gca,'xtick',[-0.2,-0.1, 0, 0.1, 0.2],'xticklabel',{'-2w','-w','0','w','2w'},'ytick',[]... | ||
,'FontSize', 14) | ,'FontSize', 14) | ||
| − | |||
% Lowpass filter to retrieve original signal | % Lowpass filter to retrieve original signal | ||
| Line 261: | Line 255: | ||
set(gca,'xtick',[-0.2,-0.1, 0, 0.1, 0.2],'xticklabel',{'-2w','-w','0','w','2w'},'ytick',[]... | set(gca,'xtick',[-0.2,-0.1, 0, 0.1, 0.2],'xticklabel',{'-2w','-w','0','w','2w'},'ytick',[]... | ||
,'FontSize', 14) | ,'FontSize', 14) | ||
| − | |||
</pre> | </pre> | ||
| + | |||
==References== | ==References== | ||
<references /> | <references /> | ||
{{Template:20.309 bottom}} | {{Template:20.309 bottom}} | ||
Revision as of 22:14, 24 January 2017
Lock-in signal detection
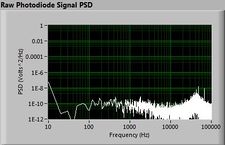
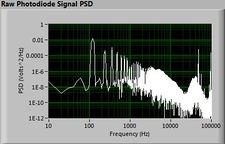
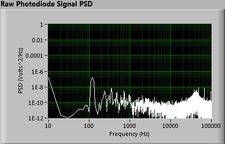
The plots below show typical power spectral density measurements of noise in the 20.309 lab — a mix of optical and electronic noise. Fluorescent lighting creates a very strong technical noise at 120 Hz and harmonics. The lock-in technique used in the DNA lab involves modulating the blue light that excites the fluorescent dye in the sample to move the signal spectrum from the riotous low frequency realm to a calmer range of frequencies. Lock-in signal detection provides some immunity to the very noisy lab environment.
Noise in lab the may look different than in previous semesters, so it may be useful to measure the noise spectrum before settling on a modulation frequency for your lock-in amplifier.
| |
||
| |
|
|
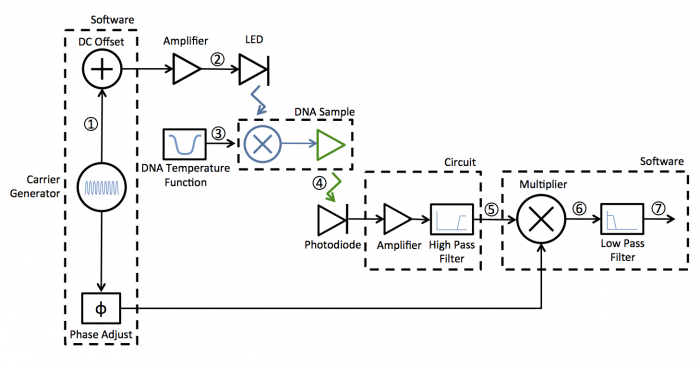
A block diagram of the lock-in scheme is shown below. Implementing lock-in detection requires the ability to modulate the LED output with a carrier frequency and requires signal processing (in software in our case) to recover the non-modulated signal. In your case the non-modulated signal is the fraction of dsDNA versus time. To support these functions, the LED circuit must be modified to include a feedback brightness controller. In addition, the photodiode amplifier must be modified to accommodate the change in signal frequency range. In the control software, you will need to choose the LED modulation frequency as well as all filter frequencies to support your design.
MATLAB Simulation
The following Matlab code was used to generate the figures above.
close all
clear all
clc
% Simulation of different steps in the lock-in amplifier
%
% Requires DnaFraction.m as a function, as documented in DNA Melting:
% Simulating DNA Melting - Basics wiki page. Steps correspond with block
% diagram shown in Understanding the Lock-in Amplifier wiki page.
%
% First, we generate a signal for DNA melting by specifying ideal
% conditions, creating a time vector, and modeling temperature.
simulationLength = 300; % 5 min sim for each melting and cooling
sampleRate = 1; % 1 Hz sample rate
sim1 = struct(); % create empty struct for sim results
sim1.concentration = 1E-6; % 1 micromolar concentration
sim1.deltaS = -184; % cal / (mole-K)
sim1.deltaH = -71E3; % cal / mole
sim1.initialTemperature = 293; % approx. room temperature
sim1.finalTemperature = 363; % 363 K = 90 C
sim1.time = 0:(1./sampleRate):(simulationLength); % create time vector (units: seconds)
sim1.coolingConstant = 200;
% use exp function to model heating and cooling
sim1.temperature = ... % Melting
(sim1.initialTemperature - sim1.finalTemperature) .* ... % T_i - T_f *
exp(-sim1.time ./ sim1.coolingConstant) + ... % e^(-t/tau) +
sim1.finalTemperature; % T_f
sim2 = struct();
sim2.initialTemperature = sim1.temperature(end);
sim2.finalTemperature = sim1.initialTemperature; % Cooling
sim2.time = (simulationLength+sampleRate):(1./sampleRate):(2*simulationLength+sampleRate);
sim2.temperature = ...
(sim2.initialTemperature - sim2.finalTemperature) .* ... % T_i - T_f *
exp(-sim1.time ./ sim1.coolingConstant) + ... % e^(-t/tau) +
sim2.finalTemperature; % T_f
sim1.time = [sim1.time,sim2.time];
sim1.temperature = [sim1.temperature,sim2.temperature];
% Signal based on simulation
dnaFractionMelting = DnaFraction(sim1.concentration, sim1.temperature,...
sim1.deltaS, sim1.deltaH);
% Model room noise using MATLAB function to generate Brown noise.
% The 'Understanding the lock-in amplifier' wiki page shows examples of
% actual noise in the room.
coloredNoiseGenerator = dsp.ColoredNoise(2,length(dnaFractionMelting),1);
brownNoise = step(coloredNoiseGenerator);
brownNoiseNorm = (brownNoise-min(brownNoise))/(max(brownNoise)-min(brownNoise))-0.4;
% First figure demonstrates that because signal and noise occupy the same
% frequencies, a bandpass filter cannot be used to remove noise from
% signal. Brown noise is added to signal and white gaussian noise is added
% on top of that.
dnaFractionNoise = awgn((dnaFractionMelting + 0.5*transpose(brownNoiseNorm)),20);
brownNoiseTransform = abs(fftshift(fft(brownNoiseNorm)));
noiseFreqs = linspace(-sampleRate/2, sampleRate/2, length(brownNoiseTransform));
signalTransform = abs(fftshift(fft(dnaFractionMelting)));
signalFreqs = linspace(-sampleRate/2, sampleRate/2, length(signalTransform));
signalNoiseTransform = abs(fftshift(fft(dnaFractionNoise)));
signalNoiseFreqs = linspace(-sampleRate/2, sampleRate/2, length(signalNoiseTransform));
figure(1)
subplot(1,2,1)
plot(sim1.time, dnaFractionNoise, 'Color', 'blue')
xlabel('Time','FontSize', 14)
ylabel('Signal','FontSize', 14)
title('Time Domain of Measured Signal','FontSize', 18)
ax = gca;
ax.XAxisLocation = 'origin';
axis([0 600 -inf inf])
set(gca,'xtick',[0],'ytick',[0,1],'FontSize', 14)
hold on
plot(sim1.time, dnaFractionMelting, '--', 'LineWidth', 2,'Color','red')
hold off
l=legend('Measured Signal with Noise (4)','Original Input Signal (3)');
set(l,'FontSize',13);
subplot(1,2,2)
plot(signalFreqs, signalTransform,'--', 'LineWidth', 1.5,'Color','red')
hold on
plot(signalNoiseFreqs, signalNoiseTransform,'Color', 'blue')
title('Frequency Domain of Measured Signal','FontSize', 18)
axis([-0.2 .2 0 inf])
xlabel('Frequency (Hz)','FontSize', 14)
ylabel('Amplitude','FontSize', 14)
set(gca,'xtick',[0],'ytick',[0],'FontSize', 14)
hold off
% Second figure shows the carrier frequency. This is the frequency (w) that we
% will move the signal to separate it from the noise. The carrier frequency
% is offset in order to prevent giving negative values to the LED.
carrierFreq =cos(2*pi*0.1*sim1.time);
carrierFreqOffset = carrierFreq + 1; % DC Offset
figure(2)
subplot(1,2,1)
plot(sim1.time,carrierFreqOffset,'Color','blue')
title('Time Domain of Carrier Frequency','FontSize', 18)
xlabel('Time','FontSize', 14)
ylabel('Signal','FontSize', 14)
ax.XAxisLocation = 'origin';
set(gca,'xtick',[],'ytick',[0],'FontSize', 14)
axis([0 600 -inf inf])
carrierTransform = abs(fftshift(fft(carrierFreqOffset)));
freqSpaceCarrier = linspace(-sampleRate/2, sampleRate/2, length(carrierTransform));
subplot(1,2,2)
plot(freqSpaceCarrier,carrierTransform, 'Color','blue')
title('Frequency Domain of Carrier Frequency','FontSize', 14)
xlabel('Frequency (Hz)','FontSize', 14)
ylabel('Amplitude','FontSize', 14)
set(gca,'xtick',[-0.2,-0.1, 0, 0.1, 0.2],'xticklabel',{'-2w','-w','0','w','2w'},'ytick',[]...
,'FontSize', 14)
axis([-0.3 .3 1E-1 inf])
% The third figure demonstrates what happens when we modulate the the
% signal to the carrier frequency by multiplying the two. The noise is
% not time varying, so the amplitude spectrum of noise will remain about
% zero, while the signal will be moved to the carrier frequency.
modulatedSignal = (carrierFreq).*dnaFractionMelting;
modulatedSigNoise = modulatedSignal + 0.3*transpose(brownNoiseNorm);
figure(3)
subplot(1,2,1)
plot(sim1.time, modulatedSigNoise, 'b' )
title('Time Domain of Modulated Signal','FontSize', 18)
xlabel('Time','FontSize', 14)
ylabel('Signal','FontSize', 14)
ax = gca;
ax.XAxisLocation = 'origin';
set(gca,'xtick',[0],'ytick',[-1,0,1],'FontSize', 14)
axis([0 600 -inf inf])
hold on
plot(sim1.time, dnaFractionMelting,'--', 'LineWidth', 2,'Color','red' )
hold off
l=legend('Modulated Signal (5)','Original Input Signal (3)');
set(l,'FontSize',13);
modulatedSigNoiseFFT = abs(fftshift(fft(modulatedSigNoise)));
modFreq = linspace(-sampleRate/2, sampleRate/2, length(modulatedSigNoiseFFT));
subplot(1,2,2)
plot(modFreq,modulatedSigNoiseFFT,'b')
title('Frequency Domain of Modulated Signal','FontSize', 18)
xlabel('Frequency (Hz)','FontSize', 14)
ylabel('Amplitude','FontSize', 14)
axis([-0.3 .3 0 inf])
set(gca,'xtick',[-0.2,-0.1, 0, 0.1, 0.2],'xticklabel',{'-2w','-w','0','w','2w'},'ytick',[]...
,'FontSize', 14)
% The fourth figure demonstrates what happens when you multiply the
% modulated signal by the reference frequency, which is at the same
% frequency as the original carrier frequency. The signal has now moved
% back to the origin, and the noise is moved to the carrier frequency.
figure(4)
subplot(1,2,1)
multipliedSignal = modulatedSigNoise .* carrierFreq;
plot(sim1.time, multipliedSignal,'b')
title('Time Domain Domain of Multiplied Signal','FontSize', 18)
xlabel('Time','FontSize', 14)
ylabel('Signal','FontSize', 14)
set(gca,'xtick',[0],'ytick',[0,1],'FontSize', 14)
axis([0 inf 0 inf])
hold on
plot(sim1.time, dnaFractionMelting,'--', 'Color','red', 'LineWidth', 2)
hold off
l=legend('Multiplied Signal (6)', 'Original Input Signal (3)');
set(l,'FontSize',13);
multipliedSigFFT = abs(fftshift(fft(multipliedSignal)));
multFreq = linspace(-sampleRate/2, sampleRate/2, length(multipliedSigFFT));
subplot(1,2,2)
plot(multFreq,multipliedSigFFT,'b')
title('Transform of Multiplied Signal','FontSize', 14)
xlabel('Frequency (Hz)','FontSize', 14)
ylabel('Amplitude','FontSize', 14)
axis([-0.3 .3 0 inf])
set(gca,'xtick',[-0.2,-0.1, 0, 0.1, 0.2],'xticklabel',{'-2w','-w','0','w','2w'},'ytick',[]...
,'FontSize', 14)
% Lowpass filter to retrieve original signal
lowpassfilter =fir1(60, 0.02/0.5);
y = conv( multipliedSignal, lowpassfilter,'valid');
t2 = linspace(0,2*simulationLength+sampleRate,length(y));
figure(5)
subplot(1,2,1)
plot(t2,y,'b')
title('Time Domain of Output Signal','FontSize', 18)
xlabel('Time','FontSize', 14)
ylabel('Signal','FontSize', 14)
set(gca,'xtick',[0],'ytick',[0,1],'FontSize', 14)
axis([0 inf 0 0.6])
hold on
plot(sim1.time,0.5*dnaFractionMelting,'--','Color','r', 'LineWidth',2)
hold off
l=legend('Output Signal (7)', 'Original Input Signal (3)');
set(l,'FontSize',13);
outputFFT = abs(fftshift(fft(y)));
outputFreq = linspace(-sampleRate/2, sampleRate/2, length(outputFFT));
subplot(1,2,2)
plot(outputFreq,outputFFT,'b')
title('Frequency Domain of Output Signal','FontSize', 18)
xlabel('Frequency (Hz)','FontSize', 14)
ylabel('Amplitude','FontSize', 14)
axis([-0.3 .3 0 inf])
set(gca,'xtick',[-0.2,-0.1, 0, 0.1, 0.2],'xticklabel',{'-2w','-w','0','w','2w'},'ytick',[]...
,'FontSize', 14)
References