|
|
| Line 10: |
Line 10: |
| | |} | | |} |
| | | | |
| − | =Introduction= | + | ==Introduction== |
| | | | |
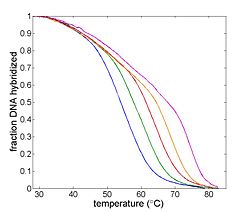
| | [[Image:Example Melting Curve.jpg|thumb|right|250px|Example DNA melting curves showing the effect of varying ionic strength. The data has been filtered to reduce noise.]] | | [[Image:Example Melting Curve.jpg|thumb|right|250px|Example DNA melting curves showing the effect of varying ionic strength. The data has been filtered to reduce noise.]] |
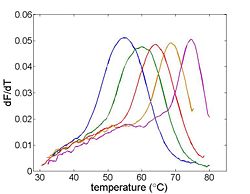
| | [[Image:Example Melting Curve Derivative.jpg|thumb|right|250px|Differentiating the melting curve simplifies finding T<sub>m</sub>.]] | | [[Image:Example Melting Curve Derivative.jpg|thumb|right|250px|Differentiating the melting curve simplifies finding T<sub>m</sub>.]] |
| − | In this lab, you will measure the melting temperature of several DNA samples to determine the effect of sequence length, ionic strength and complementarity. A common application of this technique exploits the length dependence of DNA melting temperatures to examine PCR products in order to determine whether a desired sequence was successfully amplified. | + | In this lab, you will build an instrument to measure the thermodynamic properties of DNA samples to determine the effect of sequence length, ionic strength and complementarity. A common application of this technique exploits the length dependence of DNA melting temperatures to examine PCR products in order to determine whether a desired sequence was successfully amplified. |
| | | | |
| − | The measurement technique utilizes a fluorescent dye that binds preferentially to double stranded DNA (dsDNA) <ref name="SYBR Green Paper">[http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=484200 Zipper H, Brunner H, Bernhagen J, Vitzthum F. Investigations on DNA intercalation and surface binding by SYBR Green I, its structure determination and methodological implications. ''Nucleic Acids Res.'' 2004;'''32''':e103–10.]</ref>. This characteristic of the dye allows the relative concentration of dsDNA to be determined by measuring the intensity of fluorescent light emitted by an excited sample. | + | The measurement technique utilizes a fluorescent dye that binds preferentially to double stranded DNA (dsDNA). This characteristic of the dye allows the relative concentration of dsDNA to be determined by measuring the intensity of fluorescent light given off by an excited sample. |
| | | | |
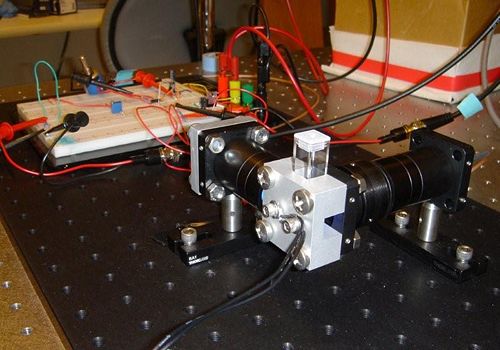
| − | The goal of your time in the lab will be to measure fluorescence intensity versus temperature for each of three samples over a range of about 75 C°, from room temperature to 95 °C. This will provide a basis for estimating the melting temperature, T<sub>m</sub>, of each sample. (T<sub>m</sub> is defined as the temperature where half of the DNA in the sample remains hybridized.) | + | The DNA melting apparatus you will construct consists of five major subsystems: excitation, fluorescence measurement, heating, temperature sensing, and data acquisition. Major components of these subsystems include an LED, a photodiode and amplifier, a thermoelectric cooler (TEC), a resistance temperature detector (RTD), and a PC data acquisition system. |
| | | | |
| − | You will measure samples of both known and unknown composition. Your samples will fall into one of several categories, each containing three different lengths, complementarities (complete match, single mismatch, or complete mismatch), or salt concentrations. You will compare the data you gather to theoretical models and to other lab groups with the same sample types, and you will attempt to identify an unknown sample. One category will be randomly assigned to each group.
| + | The goal of your time in the lab will be to measure fluorescence intensity versus temperature for each of the samples over a range of about 90°C to room temperature. This will provide a basis for estimating the melting temperature, T<sub>m</sub> of each sample. (T<sub>m</sub> is defined as the temperature where half of the DNA in the sample remains hybridized.) |
| | | | |
| − | For more information about this lab, such as the fundamentals of DNA melting and fluorescent reporters involved, plus applications such as PCR, review the links to websites, videos, and academic papers found here: [[Resource list:DNA melting and PCR]]
| + | You will measure samples of both known and unknown composition. THe samples may vary in length, complementarity (complete match, single mismatch, or complete mismatch), or salt concentration. You will compare the data you gather to theoretical models and you will attempt to identify unknown samples. |
| | | | |
| − | ===Overview of the apparatus=== | + | ===Roadmap === |
| | | | |
| − | The DNA melting apparatus you will construct consists of five major subsystems:
| + | #Build an optical system containing the LED, heating block, sample, photodiode, filters, and lenses. |
| | + | #Hook up a three terminal voltage regulator to create an electrical power supply for the LED. |
| | + | #Build, test, and calibrate the temperature-sensing circuit. |
| | + | #Build an amplification/offset circuit for the DNA fluorescence signal. |
| | + | #Troubleshoot and optimize your system. |
| | + | #Heat a samples of DNA with SYBR Green dye and record DNA melting curves both as the samples heat up, and as they cool. |
| | + | #Analyze the data: For your known samples, compare your observations to theoretical models. Identify your unknown samples. |
| | | | |
| − | #optical excitation,
| + | ==Parts of the lab manual== |
| − | #optical emission measurement,
| + | |
| − | #heating and cooling,
| + | |
| − | #temperature sensing, and
| + | |
| − | #electronic data acquisition.
| + | |
| | | | |
| − | Major components of these subsystems include, respectively, an [http://en.wikipedia.org/wiki/LED LED], the DNA itself, which is tagged with [http://en.wikipedia.org/wiki/SYBR_Green_I SYBR Green I], a [http://en.wikipedia.org/wiki/Photodiode photodiode] and [http://en.wikipedia.org/wiki/Amplifier amplifiers], a thermoelectric heater/cooler (or TEC, also called a Peltier cooler/heater because it employs the [http://en.wikipedia.org/wiki/Thermoelectric_cooling Peltier effect>], a [http://en.wikipedia.org/wiki/Resistance_thermometer resistance temperature detector (RTD)], and a PC [http://en.wikipedia.org/wiki/Data_acquisition#DAQ_hardware data acquisition system (DAQ)].
| + | *[[DNA Melting Part 1: Measuring Temperature and Fluorescence]] |
| | + | *[[DNA Melting Report Requirements for Part 1]] |
| | + | *[[DNA Melting Part 2: Lock-in Amplifier and Temperature Control]] |
| | + | *[[Matlab:Simulating DNA Melting]] |
| | + | *[[Matlab:DNA Melting Data Analysis]] |
| | + | *[[DNA Melting Report Requirements for Part 2]] |
| | | | |
| − | SYBR Green I is a fluorescent dye with peak sensitivity to blue light just below 500 nm, and the dye emits green light with an emission peak at about 520 nm.<ref name="SYBR Green Datasheet">[http://www.sigmaaldrich.com/sigma-aldrich/datasheet/s9430dat.pdf SYBR Green I online datasheet]</ref> You are encouraged to look at the spectrum [http://www.invitrogen.com/site/us/en/home/support/Research-Tools/Fluorescence-SpectraViewer.html here]. Once you have your sample you will be able to easily observe this behavior –- a room-temperature sample of dsDNA and SYBR Green I looks yellow from the combination of blue excitation and green fluorescence. At higher temperatures, when there is no dsDNA to bind the dye, the sample will appear blue or clear. In order to produce a melting curve, your apparatus will measure the amount of fluorescent emission from the SYBR Green I dye over a range of temperatures.
| + | ==Suggested readings and references== |
| | | | |
| − | Fluorescence intensity will be measured by the photodiode (and associated optical system to gather more light and concentrate it on the photodiode). Temperature will be measured by the RTD, which is a kind of resistor that predictably varies in value with temperature. The sample will be heated by the TEC. The DAQ and PC will record the signals from the photodiode and RTD. These signals must then be processed to convert them from raw voltage into temperature and fractional dsDNA concentration, ''f''. The melting temperature, ''T<sub>m</sub>'', can be estimated from a plot of ''f'' vs. ''T<sub>m</sub>'' of ''f'' vs. ''dT<sub>m</sub>/dt'' (T<sub>m</sub> differentiated with respect to time).
| + | Zipper H, Brunner H, Bernhagen J, Vitzthum F. Investigations on DNA intercalation and surface binding by SYBR Green I, its structure determination and methodological implications. ''Nucleic Acids Res.'' 2004;'''32''':e103–10.<ref name="SYBR Green Paper">[http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=484200 Zipper H, Brunner H, Bernhagen J, Vitzthum F. Investigations on DNA intercalation and surface binding by SYBR Green I, its structure determination and methodological implications. ''Nucleic Acids Res.'' 2004;'''32''':e103–10.]</ref> |
| | | | |
| − | <center>[[Image:DNA Melting Block Diagram.jpg|frame|The DNA melting apparatus includes excitation, fluorescence measurement, temperature sensing, and data acquisition functions.]]</center>
| + | A more complete [[Resource list:DNA melting and PCR|DNA melting and PCR resource list]] is available on this wiki. Please improve the page by adding relevant, high-quality sources. |
| − | | + | |
| − | As shown in the diagram, an aluminum heating block holds a cuvette containing the sample under test. The sample contains a combination of DNA, SYBR Green I, and a salt (either KCl or NaCl, it doesn't matter which). In addition to being a convenient holder, the block transfers heat to the cuvette and it's sample, while also allowing the block's temperature to be measured without affecting the block temperature. (In your report, you should tell us about the practical problems with this approach and ways to improve it. Better yet, improve it '''then''' tell us about it.)
| + | |
| − | | + | |
| − | Blue light from an LED illuminates one side of the cuvette. A 470 nm bandpass optical filter shapes the output spectrum of the LED so that only the desired wavelengths of light fall on the sample.
| + | |
| − | | + | |
| − | A photodiode placed at 90 degrees to the LED source detects the green light emitted by SYBR Green I bound to the minor groove of dsDNA. The photodiode is placed after a 515 nm long pass optical filter to ensure that only green and longer wavelength light is detected. (In your report, can you describe how do we ensure that only the fluorescent light given off by the sample falls on the photodiode?)
| + | |
| − | | + | |
| − | Since photodiodes produce only a very small amount of current, it will be necessary to build a very high gain amplifier to produce a signal that is measurable by the PC data acquisition (DAQ) cards. Photodiode amplifiers are particularly challenging because many of the non-ideal characteristics of op amps become apparent at high gain. For this reason it is suggested that you build a two-stage amplifier. And furthermore it is suggested that your first stage be a transimpedence amplifier like the one shown in the figure. (Can you comment on why a transimpedence amplifier is better for photodiode output amplification?)
| + | |
| − | | + | |
| − | An RTD attached to the heating block and wired to a voltage divider provides an indication of temperature. The temperature of the heating block will be a proxy for the sample temperature. It is not a perfect proxy, however, and you will need to determine the transfer function between the temperature you measure on the outside of the block, and the temperature inside the cuvette, say, in the middle of your DNA sample. Refer to your lecture notes and see your Instructors or TA for help with this.
| + | |
| − | | + | |
| − | The DAQ digitizes the amplified photodiode and RTD signals. A LabVIEW virtual instrument (VI) is provided that will control the heating and cooling of your DNA heating block and record the RTD and photodiode signals over time so that data from the DNA melting VI can be saved to a file. The file can be loaded into Matlab for analysis, or if you prefer, Python or equivalent.
| + | |
| | | | |
| | ===Objectives and learning goals=== | | ===Objectives and learning goals=== |
| | | | |
| − | *Build an optical system for exciting the sample with blue light and gathering the green fluorescence output on the photodiode. | + | *Build an optical system for exciting the sample with blue light and gathering the fluorescence output on the photodiode. |
| | + | *Measure light intensity with a photodiode. |
| | *Build a heating system to reliably heat and cool your sample. | | *Build a heating system to reliably heat and cool your sample. |
| − | *Measure light intensity with a photodiode.
| |
| | *Measure temperature with an RTD and an appropriate transfer function. | | *Measure temperature with an RTD and an appropriate transfer function. |
| | *Implement a high gain transimpedance amplifier. | | *Implement a high gain transimpedance amplifier. |
| − | *Record dsDNA fluorescence versus temperature for several samples. | + | *Use a lock-in amplifier to reduce noise. |
| | + | *Record dsDNA concentration versus temperature curves for several samples. |
| | *Analyze the data to find the dsDNA fraction as a function of temperature. | | *Analyze the data to find the dsDNA fraction as a function of temperature. |
| | *Estimate T<sub>m</sub> from your data. | | *Estimate T<sub>m</sub> from your data. |
| | *Compare the measured curves with theoretical models. | | *Compare the measured curves with theoretical models. |
| | *Identify unknown DNA samples. | | *Identify unknown DNA samples. |
| − |
| |
| − | =Lab procedure=
| |
| − |
| |
| − |
| |
| − |
| |
| − | ===Optical system===
| |
| − |
| |
| − | <center>[[Image:DNA Melting Optical System Diagram.jpg|700 px|thumb|center|DNA Melting Optical System Diagram]]</center>
| |
| − |
| |
| − | The optical system consists of an LED, excitation filter, sample cuvette, heating block, emission filter, photodiode, optional lenses, and associated mounting hardware. Construct your system on an [http://www.thorlabs.com/thorProduct.cfm?partNumber=MB1224 optical breadboard]. The breadboard has a grid of tapped holes for mounting all kinds of optical and mechanical hardware. [http://www.thorlabs.com ThorLabs] manufactures most of the hardware stocked in the lab. A few of the components you will certainly use include: [http://www.thorlabs.com/newgrouppage9.cfm?objectgroup_id=1266 1/2" diameter posts], [http://www.thorlabs.com/thorProduct.cfm?partNumber=CP02 CP02 cage plates], and [http://www.thorlabs.com/NewGroupPage9.cfm?ObjectGroup_ID=1521 1” diameter lens tubes].
| |
| − |
| |
| − | Use [http://www.thorlabs.com/thorProduct.cfm?partNumber=RLA0300 optical rails] and [http://www.thorlabs.com/thorProduct.cfm?partNumber=RC1 rail carriers] or [http://www.thorlabs.com/NewGroupPage9.cfm?ObjectGroup_ID=47 optical bases] to mount 1/2” posts on the breadboard. [http://www.thorlabs.com/thorProduct.cfm?partNumber=RA90 RA90] right angle post clamps and [http://www.thorlabs.com/thorProduct.cfm?partNumber=PH2-ST post holders]can also be useful.
| |
| − |
| |
| − | There are a variety of ways to construct the apparatus. A good design will be compact, stable, and simple. It is a good idea to shield the optical system from ambient light, so a small footprint will be advantageous.
| |
| − |
| |
| − | ====Illumination====
| |
| − |
| |
| − | You will use a high-brightness blue LED manufactured by LedEngin, Inc. to excite the sample ([http://www.ledengin.com/led_products.htm datasheet]). This LED has a dominant wavelength of 465 nm. A Chroma Technology [http://web.mit.edu/~20.309/Students/Datasheets/Chroma%20D470-40.pdf D470/40] filter eliminates unwanted parts of the spectrum that might interfere with detection of the fluorescence signal, such as the very wavelengths you want to measure as fluorescent output. The filters have exposed, delicate coatings and must be [http://www.chroma.com/index.php?option=com_content&task=view&id=56&Itemid=65 handled carefully]. In addition, the filter [http://www.chroma.com/support/ordering-information/orientation-instructions works better in one direction than the other].
| |
| − |
| |
| − | Light from the LED diverges in a cone with an angle of about 100 degrees. It may be advantageous to include optics in your system to concentrate light from the LED on to the sample.
| |
| − |
| |
| − | ====Fluorescence detection====
| |
| − |
| |
| − | The [http://www.thorlabs.com/Thorcat/8700/8770-D02.pdf SM05PD1A] photodiode is mounted in a short tube with SM05 threads. Use a [http://www.thorlabs.com/thorProduct.cfm?partNumber=SM1A6 SM1A6] adapter to mount the photodiode in a CP02 cage plate or lens tube. Mount the photodiode assembly to the breadboard at 90 degrees to the LED. Build a system to hold the Chroma [http://web.mit.edu/~20.309/Students/Datasheets/Chroma%20E515lpv2.pdf E515LPV2] emission filter in front of the photodiode. You can use lenses to focus light from the sample on to the detector to improve performance, if you like.
| |
| − |
| |
| − | Do not connect the sample block to any other pieces of metal — this will draw off heat and affect the thermal cycler's performance. If you want to link the sample block to the fluorescence detection arm, use hexagonal plastic standoffs to connect two cage rails without forming a thermal linkage.
| |
| − |
| |
| − | ===Electrical System===
| |
| − | ====Temperature detection====
| |
| − |
| |
| − | [[Image:rtd2.png|right|thumb|250px|Temperature Detection Circuit]]
| |
| − |
| |
| − | The electrical resistance of most materials varies with temperature. RTDs are a special resistors (usually made out of platinum) that exhibit a nearly linear change in its value with temperature. An RTD may be used to accurately measure temperature by including it as an element in a voltage divider. As the resistance of the RTD changes, so will the voltage across it.
| |
| − |
| |
| − | A [http://www.ussensor.com/prod_rtds_thin_film.htm PPG102A1 RTD] has been pre-mounted to the DNA heating block. This RTD has a nominal resistance of 1 KΩ and its value increases with temperature. Note that the maximum current carrying capacity of this device is 1 ma. Hook up the RTD in a voltage divider. Make sure the divider has no more than 1 mA flowing through it. Use freeze spray or heat the block on the warmer to test the circuit.
| |
| − |
| |
| − | The Labview VI calculates the temperature of the heating block based on the voltage across the RTD. This calculation assumes the use of +15V and 15KΩ. The diagram to the right shows the suggested circuit. The capacitor across the RTD implements a low pass filter. The purpose of this filter is to eliminate noise in your temperature reading. Consider the following points when choosing a capacitor value: What is the frequency range that you want filtered? The time constant of the filter is determined by the two resistors in parallel with the capacitor. What is the time constant of your filter? If it is too large compared to the rate of change of temperature, it could cause a delay in your temperature reading.
| |
| − |
| |
| − | ====Heating control====
| |
| − |
| |
| − | We will use a thermoelectic cooler (TEC) to heat the sample in this lab. A TEC is a device that becomes warm on side and cold on the other side when a DC voltage is applied across it. Before heating the sample, make sure that the TEC makes good thermal contact with the heating block. If there is not good thermal contact, hot spots will develop damaging the TEC. To ensure good contact, use the MINIMUM amount of thermal grease, and make sure that the heating block lies flat on the TEC. Wooden sticks are available in the DNA Melting drawer (far right of the prep bench) to help you apply an extremely thin schmear.
| |
| − |
| |
| − | To control the TEC we will use a digital output from the Labview VI and a solid state relay. The relay is an electronic switch with four pins. Two pins are for the control voltage and the other pins are the two sides of the switch. When the control voltage is "high", the switch is turned "on" (this means the switch is closed = the two pins are connected together). The Labview VI provides a digital output from your DAQ that will serve as the control voltage. Be sure to see your TA for a TEC Safety Circuit that you must add in series with the DAQ digital output.
| |
| − |
| |
| − | The LabView VI allows you to set the temperature and heating rate that you want. The VI compares the desired heating rate to the current heating rate (determined by the time derivative of the voltage across the RTD). The VI drives the relay with a square wave of constant period and variable duty cycle (= % of the time the square wave is "high" vs. "low"). If the current heating rate is too slow, then the duty cycle is increased (up to a limit of 0.5). If the current heating rate is too fast, then the duty cycle is decreased (down to a limit of 0.001).
| |
| − |
| |
| − | The diagram below is the suggested schematic for the lab. The positive side of the TEC is always connected to +12V through the 250W computer power supply. The supply is capable of sourcing 5 A at 12 V. The negative side of the TEC is connected to the first pin of the relay (one side of the switch). The other side of the relay is connected to ground, which is also connected to the ground of the 250W power supply.
| |
| − |
| |
| − | When the control voltage is high, the switch is closed and the negative side of the TEC is connected to ground. Now that the negative side of the TEC is connected to ground, there is a DC voltage across the TEC and current flowing through it, which will cause the TEC to heat up. When the switch is open, the TEC does not have a path to ground and the TEC will not heat up.
| |
| − |
| |
| − | <center>[[Image:Heatcontrol.png|500 px|thumb|center|Heating Control Circuit (note that the '''black''' side of the relay is the "front" as shown in this figure)]]</center>
| |
| − |
| |
| − | ====LED driver====
| |
| − |
| |
| − | [[Image:Led2.png|right|thumb|250px|LED Driving Circuit.]]
| |
| − |
| |
| − | The LED used in this lab can consume 1.5 Amps of current when maximally on, but this is MUCH more than you need. You will need only roughly 100 mA, but even this low current is 50x what the [[20.309:DAQ System|20.309 USB-6212]] DAQ system is capable of driving. It is possible to drive an LED with a voltage source; however, the steepness of a diode's I-V curve results in large current swings in current for small changes in supply voltage. LED brightness is proportional to current, so a current source tends to provide a more stable light output.
| |
| − |
| |
| − | To amplify the current drive to the LED, you will be using a Darlington Transistor. The transistor is a three terminal device, consisting of a Base, Emitter, and Collector, which should be wired into the circuit shown below. The base contact of the BJT delivers a small amount of current from the DAQ, which causes the device to conduct a much larger current through the LED.
| |
| − |
| |
| − | In this lab you will linearize the behavior of the LED and transistor by using the op amp circuit we discussed in class. Recall that this is simply a buffer implementation, with gain of 1, where the feedback voltage is directly proportional to the LED current. Note that the voltage to the BJT and LED setup comes from the fixed 5V supply, whereas the op amp operates on +/– VCC (+/-15V).
| |
| − |
| |
| − | Double check your wiring before connecting the LED. The LED can be damaged by excessive current. Remember the rule of thumb: if you can't keep your finger on a component indefinitely, it is too hot. Use a larger feedback resistor to keep the electronics cooler (at the expense of light output).
| |
| − |
| |
| − | If the LED is too bright the sample will become photobleached very quickly. You will be able to set the amplitude and offset of the the sinusoid driving the LED. This is set in the Labview VI. Set the offset so that the sinusoid is always positive (i.e. if the amplitude is 100mV then the offset should be at least 100mV). To reduce switching noise, you can set the offset just slightly higher than it needs to be (e.g. 105mV), so that the current never goes all the way to zero.
| |
| − |
| |
| − | ====Photodiode amplifier====
| |
| − | Excited SYBR Green I molecules emit photons in all directions, some of which pass through the emission filter and strike the photodiode. This generates a tiny current, which must be converted to a voltage and amplified to a level that can be conveniently measured. From past experience, the photodiode output current will be on the order of 15 nA. (Can you calculate it from first principles?)
| |
| − |
| |
| − | =====Amplification circuit=====
| |
| − |
| |
| − | The NI-DAQ interface card used in lab can digitize a voltage between positive and negative 10 Volts. A first approach to providing this voltage swing at the digitizer inputs was demonstrated in the homework: a transimpedance amplifier (sometimes called a current-to-voltage converter) with a gain of approximately <math>10^6-10^7</math> V/A will produce an output that is in this range, with some room for a stronger sample response, given a 15 nA input. (Why not simply use a resistor, and omit the op-amp?)
| |
| − | Implementing the photodiode amplifier in a single stage presents a number of challenges, however, as many of the non-ideal behaviors of op amps become apparent at such high gain values. You will be implementing a three-stage photodiode amplifier, as shown in the figure below. You are not limited to the topologies shown here; you may use others for one of the stages, such as the transimpedance amplifier from your homework. If you have another in mind, feel free to discuss with your TA and/or Instructors. Your job as circuit designers will be to at least fill in the appropriate filtering capacitor and feedback resistor values, in the topology shown here, in order to achieve the desired gain at the output.
| |
| − |
| |
| − | Think carefully about how you want to distribute the gain of the system across the three stages: how does feedback affect the noise of the amplifier? What are the dominant sources of noise in the DNA melting setup, which ones do you have most control over, and how will you control for them? Applying your knowledge of feedback and transfer functions, you should be able to determine the optimal gain configuration of each stage for lowest output noise.
| |
| − |
| |
| − | <center>[[Image:DNA_Amplifier.jpg|700 px|thumb|center|Multi-stage photodiode amplification circuit]]</center>
| |
| − |
| |
| − | Even with these optimizations, it will be important to keep your wiring short and neat. The amplifier and wiring will also be susceptible to physical movement, so prevent things from getting bumped during experimental runs. Select an op amp that has as low input bias current as possible. (Why?) The DNA melting lab is stocked with a variety of op amps – you should familiarize yourself with the data sheets provided with these devices when making your selections. Finally, a note about resistors and capacitors: recall that resistors introduce thermal noise, while capacitors do not. Therefore, the optimized system will use as low resistor values as are reasonable in filters and feedback paths.
| |
| − |
| |
| − | ===Practical matters===
| |
| − |
| |
| − | <blockquote>
| |
| − | <div>
| |
| − | ''As with most amplifiers, care should be taken with lead dress, component placement and supply decoupling in order to ensure stability.''
| |
| − |
| |
| − | ''—[http://www.ortodoxism.ro/datasheets/nationalsemiconductor/DS005655.PDF LF411 Datasheet]''
| |
| − |
| |
| − | ''In theory, there is no difference between theory and practice. But, in practice, there is.''
| |
| − |
| |
| − | ''—[http://en.wikiquote.org/wiki/Jan_L._A._van_de_Snepscheut Jan L. A. van de Snepscheut]/[http://en.wikiquote.org/wiki/Yogi_Berra Yogi Berra]/and or [http://c2.com/cgi/wiki?DifferenceBetweenTheoryAndPractice Chuck Reid]''
| |
| − |
| |
| − | </div>
| |
| − | </blockquote>
| |
| − | The universe is rife with electrical noise. Keeping the noise out of sensitive electronic instruments requires a great deal of care. Unfortunately, electronic breadboards are a poor environment in which to construct high gain amplifiers. A few simple tricks can improve things.
| |
| − |
| |
| − | *Use power supply bypass capacitors. Connect a large capacitor between all supply voltages and ground. Large, electrolytic capacitors of at least 0.1 μF work well for this purpose. Electrolytic capacitors are polarized. Make sure to put them in the right direction (They may explode, but they will definitely start to smell bad.).
| |
| − | *Use the binding posts on the breadboard to connect your power supplies.
| |
| − | *Power Supplies: Use a seperate power supply for the op-amp circuits, LED driving circuit, and the heating control. Make sure to tie the 5V ground to +15V/-15V ground at the power supply. If you connect the grounds on the breadboard, there will be a potential difference between the grounds. This voltage difference is due to the large current swing of the LED driver flowing through the parasitic resistance of the breadboard.
| |
| − |
| |
| − | ===PC Data Acquisition System===
| |
| − |
| |
| − | {{:20.309:DAQ System}}
| |
| − |
| |
| − | ====Summary of DAQ inputs/outputs====
| |
| − |
| |
| − | {| class="wikitable"
| |
| − | |-
| |
| − | ! Signal
| |
| − | ! High Side
| |
| − | ! Low Side or Ground
| |
| − | |-
| |
| − | ! RTD
| |
| − | | AI0+
| |
| − | | AI0-
| |
| − | |-
| |
| − | ! Photodiode
| |
| − | | AI1+
| |
| − | | AI1-
| |
| − | |-
| |
| − | ! LED Driver
| |
| − | | AO0
| |
| − | | AOGND
| |
| − | |-
| |
| − | ! Heat/Cool
| |
| − | | P1.0
| |
| − | | DGND
| |
| − | |-
| |
| − | ! Heat/Cool On/Off
| |
| − | | AO1
| |
| − | | AOGND
| |
| − | |}
| |
| − |
| |
| − | ====Things to watch out for====
| |
| − |
| |
| − | *The DAQ has only one ADC (analog-to-digital converter), which is multiplexed between all of its analog inputs. The ADC has input limits of +/-10V. If one input is outside this range, it will get confused and give nonsensical values for all inputs. The upshot is, if either your temperature or fluorescence measurement is nonsensical, check to make sure the other one isn't feeding +15V into the DAQ.
| |
| − | * If you short-circuit your DAQ, it will get confused. One symptom of this is the LabView VI refusing to run (for example, wrongly complaining that the limits on the band-pass filter do not satisfy some condition). If this happens, unplug and re-plug the DAQ and use the Measurement and Automation software to check its condition.
| |
| − |
| |
| − | ====LabVIEW VI====
| |
| − |
| |
| − | The DNA Melting LabVIEW VI is located in the <code>Students/Labs/DNA Melting</code> folder of the course locker. Double click to launch the VI. (The current version is R1.0)
| |
| − |
| |
| − | Click the run arrow or select Operate->Run from the menu to start the VI. The top two charts show the digitized voltage at the RTD and diode inputs over time. Use the range settings to get a good view of the signal.
| |
| − |
| |
| − | At the end of an experimental run, use the <code>Save Data</code> button to save the most recent result in a tab-delimited file that can be read into Matlab or Excel.
| |
| − |
| |
| − | ===Debugging the apparatus===
| |
| − |
| |
| − | #Use freeze spray and the heat gun to make sure the temperature circuit is working properly.
| |
| − | #Cover and uncover the photodiode to verify operation of the fluorescence measurement system.
| |
| − | #Use a box and a piece of black cloth to shield your apparatus from ambient light. Can you measure the difference between a cuvette filed with water and one with DNA and SYBR Green?
| |
| − | #Observe every electrical signal node with the oscilloscope. Are any signals noisy? Is there a way to improve the quality of poor signals?
| |
| − | #Watch the fluorescence readout over time. Is it stable or does it drift?
| |
| − | #Is your temperature reading nonsensical? Make sure you are using 15V supply and 15kohm resistor, for your voltage supply. Also make sure that the DAQ input is placed across the RTD not the 15Kohm resistor.
| |
| − | #Is your LED being driven at the desired carrier frequency? Try turning the carrier frequency down low enough to see it turn on and off (try 5Hz). Try pointing the LED directly at the photodiode. Is the frequency of the photodiode signal what you expect?
| |
| − | #Is your sampling photobleaching quickly? Turn the amplitude of the LED signal down at the Labview VI. Try an amplitude of 0.25 V and an offset 0.50 V. Turning the amplitude down decreases signal strength, but you can gain signal strength by focusing the green fluorescent light on the photodiode with lenses.
| |
| − |
| |
| − | ==Experimental procedure==
| |
| − |
| |
| − | Samples are in limited supply, as usual, so to receive your test samples you must first confirm that your apparatus is functioning properly and that your analysis script is functional. Your group can actually get your code running at the same time as your instrument is being created. You may either generate some data from the [[Matlab:Simulating DNA melting|Simulating DNA melting]] wiki page, or you may use previous-run data that we provided on Stellar.
| |
| − |
| |
| − | To demonstrate a properly running instrument, your TA will provide a sample of 20bp DNA and will evaluate your raw data. As a first step, turn your apparatus on and measure the difference between a cool DNA sample and water. Run the DNA melting LabVIEW VI in the DNAMelting directory of the course locker (First make a copy for yourself in your directory.).
| |
| − |
| |
| − | Once your instrument is running to your (and your TA's) satisfaction, you may measure melting curves of each of the test samples. And if you have time, you can run additional experiments. For example, you could gather additional ionic strength data points.
| |
| − |
| |
| − | The DNA melting apparatus will generate the best data when both the amplifier circuit and LED have been on for a while and all the components have reached their steady state temperature. Make sure the outputs of the system are stable before you begin taking data.
| |
| − |
| |
| − | The steps for each experimental run are:
| |
| − |
| |
| − | #Open the LabView VI
| |
| − | #Set the carrier frequency, the LED amplitude, the desired temperature, and the filter cutoff frequencies
| |
| − | #Then prepare a sample and place cuvette in heating block
| |
| − | #Run the Labview: recording RTD and photodiode output during heating
| |
| − | #Switch off the heater and record RTD and photodiode output during cooling
| |
| − | #Make any modifications to the code that will help you
| |
| − |
| |
| − | ===Make a sample===
| |
| − |
| |
| − | {{Template:Safety Warning|message=SYBR Green I in DMSO is readily absorbed through skin. Synthetic oligonucleotides may be harmful by inhalation, ingestion, or skin absorption. Wear gloves when handling samples. Wear safety goggles at all times when pipetting the SYBR Green/DNA samples. Do not create aerosols. The health effects of SYBR Green I have not been thoroughly investigated. See the SYBR Green I and synthetic oligonucleotide MSDS in the course locker for more information.}}
| |
| − |
| |
| − | Pipet 500μl of DNA plus dye solution into a glass cuvette. (Ask your TA or an Instructor for one of these reusable, and expensive, cuvettes). Pipet 20μl of mineral oil on top of the sample to help prevent evaporation. Put a disposable top (available in a drawer under the bench) on the cuvette and mark it with a permanent marker. Keep the sample vertical to make sure the oil stays on top. You should be able to use the same sample for several heating/cooling cycles (What makes it go bad? What can you do to limit this effect?). Only discard it if you lose significant volume due to evaporation or your signal gets too low. If you need to leave the sample overnight, store it in the lab refrigerator. If you finish with a sample and it is still in good shape, pass it on to another group.
| |
| − |
| |
| − | ===Sample Disposal===
| |
| − |
| |
| − | {{Template:Environmental Warning|message=Discard pipette tips with DNA sample residue in the ''Biohazard Sharps'' container. Do not pour synthetic oligonucleotides or SYBR Green I down the drain. Empty the liquid into the waste container provided. Clean your glass cuvettes by flushing with alcohol in the waster container, rinsing with water at the drain, and if you wish by also soaking/rinsing with the Lysol cleaner. You can suck out residual liquid with the vacuum and drawn pipette to the left of the sink.}}
| |
| − |
| |
| − | ==Data Analysis==
| |
| − | Use Matlab or another environment of your choosing to analyze the raw data. If you prefer Python, consult [[Python:DNA melting data analysis | this page]]. In outline, the steps are:
| |
| − |
| |
| − | #Load data file produced by LabVIEW VI into Matlab
| |
| − | #Apply median filter to reduce impulse noise
| |
| − | #Apply FIR filter to reduce other sources of noise
| |
| − | #Convert RTD voltage to temperature
| |
| − | #Normalize fluorescence signal
| |
| − | #Apply transfer function to measured RTD temperature to calculate sample temperature
| |
| − | #Sort or bin the data to ensure that fluorescence versus temperature is a function
| |
| − | #Average data from multiple experimental runs, if necessary
| |
| − | #Apply correction for photobleaching, if necessary
| |
| − | #Apply correction or SYBR Green I temperature dependence, if possible
| |
| − | #Compute the finite difference Δfluorescence/Δtemperature
| |
| − | #Estimate the melting temperature
| |
| − | #Create plots
| |
| − |
| |
| − | ===Organization of data analysis script===
| |
| − | Since you will be analyzing several experimental runs in a similar way, it makes sense to save the particulars of each experimental run (such as the name of the file that the data is stored in and the KCl concentration) in an array of data structures. Matlab implements a data type called a ''cellular array''. Cellular arrays use pointy braces {} instead of parenthesis (). If <code>foo</code> is a cellular array, then <code>foo{1}</code> is the first element.
| |
| − |
| |
| − | The following code demonstrates how to create the cellular array of data structures. The code fills in the first element of a cellular array (<code>DnaMatchSampleInfo</code>):
| |
| − |
| |
| − | <pre>
| |
| − | DnaMatchSampleInfo = {} % null cellular array
| |
| − |
| |
| − | DnaMatchSampleInfo{1}.filename = 'DNA Melting Data\20bp 100mM.txt';
| |
| − | DnaMatchSampleInfo{1}.SampleName = '20 bp 100mM';
| |
| − | DnaMatchSampleInfo{1}.KclConcentration = 100;
| |
| − | DnaMatchSampleInfo{1}.DnaConcentration = 30;
| |
| − | DnaMatchSampleInfo{1}.FilterNormalizedCutoffFrequency = 0.1;
| |
| − | DnaMatchSampleInfo{1}.FilterOrder = 25;
| |
| − | DnaMatchSampleInfo{1}.NormalizationMin = 0.01;
| |
| − | DnaMatchSampleInfo{1}.NormalizationMax = 0.99;
| |
| − | DnaMatchSampleInfo{1}.TrimStart = 0;
| |
| − | DnaMatchSampleInfo{1}.TrimEnd = 0;
| |
| − |
| |
| − | ... fill in more entries here ...
| |
| − | </pre>
| |
| − |
| |
| − | After initializing <code>DnaMatchSampleInfo</code>, each element will contain a data structure with the parameters of a particular experimental run. You can create additional cellular arrays for different sets of experiments. For example, <code>DnaLengthSampleInfo</code> might contain parameters for the 20, 30, and 40 base pair samples.
| |
| − |
| |
| − | Using the information in the cellular arrays, it is easy to write a <code>for</code> loop that will process all of the data for a particular set of experiments. The function (called <code>DnaMelt</code> might look something like this:
| |
| − |
| |
| − | <pre>
| |
| − | % Function to process DNA melting data from LabVIEW VI
| |
| − | % sampleInfo is a cellular array of data structures
| |
| − | % output is a cellular array containing original data and computed values
| |
| − | function out = DnaMelt(sampleInfo)
| |
| − |
| |
| − | % initialize variables
| |
| − | out = {};
| |
| − |
| |
| − | for ii=1:length(sampleInfo)
| |
| − |
| |
| − | ... process the data ...
| |
| − |
| |
| − | end
| |
| − | </pre>
| |
| − |
| |
| − | ... process/plot specific results about whole data set...
| |
| − |
| |
| − | ===Loading a data file===
| |
| − | Use the <code>load</code> command to read in a data file.
| |
| − |
| |
| − | <pre>
| |
| − | rawData = load(sampleInfo{ii}.filename);
| |
| − | </pre>
| |
| − |
| |
| − | The first column contains RTD voltage samples and the second contains the corresponding fluorescence voltages.
| |
| − |
| |
| − | ===Converting voltages to temperature and relative fluorescence===
| |
| − | These are straightforward mathematical manipluations on the dataset using the properties of the RTD and your normalized/corrected/adjusted fluorescence signal with the equation for dsDNA fraction.
| |
| − |
| |
| − | ===Designing an FIR low pass filter===
| |
| − | The matlab commands <code>fir1</code> and <code>freqz</code> are useful for designing a low pass filter. Use <code>fir1</code> to generate the filter kernel and <code>freqz</code> to plot its frequency response. Increasing the order of the filter will make the transition between pass and stop bands sharper.
| |
| − |
| |
| − | Try plotting a few different filter designs:
| |
| − |
| |
| − | <pre>
| |
| − | freqz(fir1(30, 0.15))
| |
| − | freqz(fir1(300, 0.15))
| |
| − | </pre>
| |
| − |
| |
| − | ===Applying the FIR filter===
| |
| − | Use the <code>conv</code> command to apply the filter. But be careful how you treat the samples near the beginning and end of the signal. <code>conv</code> returns a vector of length m + n - 1, where m and n are the lenghts of the two operands. <code>conv</code> will assume that all values outside of the defined signal are zero. This will distort your signal near the beginning and end. You can handle this by pre-padding your data with made-up data (usually the initial and final values) or just chopping off the extra samples.
| |
| − |
| |
| − | The following functions may be of use:
| |
| − |
| |
| − | <pre>
| |
| − | % usage: ConvolveAndClip(kernel, data)
| |
| − | % convolves <kernel> with <data> and trims the ends of the result to length
| |
| − | % length(data) - length(kernel)
| |
| − | function out=ConvolveAndClip(kernel, data)
| |
| − |
| |
| − | temp = conv(kernel, data);
| |
| − | trim = length(kernel);
| |
| − | out = temp(trim:end-trim);
| |
| − | </pre>
| |
| − |
| |
| − | <pre>
| |
| − | % usage: PadAndConvolve(kernel, data)
| |
| − | % Pads <data> with initial and final values, convolves <data> with
| |
| − | % <kernel>, and trims the result to the same length as <data>
| |
| − | function out=PadAndConvolve(kernel, data)
| |
| − |
| |
| − | frontPaddingLength = floor(length(kernel)/2);
| |
| − | rearPaddingLength = ceil(length(kernel)/2);
| |
| − | frontPadding = data(1) * ones(frontPaddingLength, 1);
| |
| − | rearPadding = data(end) * ones(rearPaddingLength, 1);
| |
| − |
| |
| − | s = size(data);
| |
| − |
| |
| − | if(s(1) > s(2))
| |
| − | paddedData = [frontPadding;data;rearPadding];
| |
| − | else
| |
| − | paddedData = [frontPadding' data rearPadding'];
| |
| − | end
| |
| − |
| |
| − | out = ConvolveAndClip(kernel, paddedData);</pre>
| |
| − |
| |
| − | ===Ensuring that F(Temperature) is a function===
| |
| − | Because there is noise in the temperature readout, your raw data is not guaranteed to be a function. You will run into all sorts of trouble taking the finite difference later on if your data is not a function. This can be solved by either sorting the data by temperature (using Matlab's <code>sort</code> command) or binning the samples by temperature (by iterating through the data with a <code>for</code> loop.)
| |
| − |
| |
| − | Each approach has merits and disadvantages. Sorting by temperature can result in some very small ΔT values, which tend to be very noisy. Binning has the advantage of resulting in a uniformly sampled dataset — provided that there is at least one sample in each bin.
| |
| − |
| |
| − | ===Computing the finite difference===
| |
| − | The <code>diff</code> command will compute the finite difference of a discrete-time signal, which is an approximation of a continuous derivative. Remember that you need to compute ΔFluorescence/ΔTemperature, not ΔFluorescence/ΔTime. Therefore, you must divide by ΔTemperature.
| |
| − |
| |
| − | The differencing operation is particularly sensitive to noise. (What is the frequency response of a differencer?) If your derivative plots are noisy, you may be able to improve them by applying additional filtering.
| |
| − |
| |
| − | One estimate of ''T<sub>m</sub>'' is the peak value of the numerical derivative.
| |
| − |
| |
| − | ===Fitting the model===
| |
| − | In the "Simulating DNA Melting" tutorial for [[Matlab:Simulating DNA melting|Matlab]] or [[Python:Simulating DNA Melting|Python]], there is a function for computing the theoretical value of dsDNA concentration as a function of DNA concentration, temperature, ΔH° and ΔS°.
| |
| − |
| |
| − | To perform the fit, use the matlab function from the tutorial. You can use Matlab's curve fitter (<code>lsqcurvefit</code>) to estimate best-fit values for ΔH° and ΔS°.
| |
| − |
| |
| − | <code>lsqcurvefit</code> requires the fitting function to be implemented a particular way. You may find the following code excerpt, which declares a suitable function called <code>myFunction</code>, useful. It also computes the R squared value for the fit.
| |
| − |
| |
| − | <pre>
| |
| − | % Create user function for fitting
| |
| − | DnaConcentration = sampleInfo{ii}.DnaConcentration;
| |
| − | myFunction = @ (x, xdata) DnaFraction(DnaConcentration, xdata, x(1), x(2));
| |
| − |
| |
| − | % Fit data to model
| |
| − | fitOptions = optimset('TolFun',1E-30,'TolX',1E-10,'MaxFunEvals',1E4,'MaxIter',1E4);
| |
| − | [fitValues, resnorm] = lsqcurvefit(myFunction, [-150 -71E3], temperature, ...
| |
| − | fluorescence, [-inf -inf], [inf inf], fitOptions);
| |
| − | rSquared = 1 - resnorm / norm(fluorescence - mean(fluorescence))^2;
| |
| − |
| |
| − | </pre>
| |
| − |
| |
| − | ===Plot legends in Matlab===
| |
| − | Annoyingly, Matlab handles legends differently than other plot commands. The <code>hold</code> command does not apply to the <code>legend</code> command. (Direct complaints to http://www.mathworks.com/support.) The following code excerpt adds a plot legend. This code runs after the main loop. The cellular array <code>out</code> contains the results from processing. This code also places an "X" at the melting temperature.
| |
| − |
| |
| − | <pre>
| |
| − | % Compute text cell array for plot legends and plot an
| |
| − | % "X" at the estimated melting temperature
| |
| − |
| |
| − | for ii=1:length(out)
| |
| − | legendText{2 * ii - 1} = sampleInfo{ii}.SampleName;
| |
| − | legendText{2 * ii} = [sampleInfo{ii}.SampleName ' Best Fit'];
| |
| − | figure(1)
| |
| − | plot(out{ii}.maxDerivativeTemperature, out{ii}.fluorescence(out{ii}.maxDerivativeIndex),...
| |
| − | 'linewidth',2,'marker','x','markersize',18,'color',out{ii}.plotColor);
| |
| − | plot(out{ii}.maxModelDerivativeTemperature, out{ii}.fitFluorescence(...
| |
| − | out{ii}.maxModelDerivativeIndex), 'linewidth',2,'marker','x','markersize',18,'color',...
| |
| − | out{ii}.modelPlotColor);
| |
| − | figure(2)
| |
| − | plot(out{ii}.maxDerivativeTemperature, out{ii}.dFdT(out{ii}.maxDerivativeIndex),...
| |
| − | 'linewidth',2,'marker','x','markersize',18,'color',out{ii}.plotColor);
| |
| − | plot(out{ii}.maxModelDerivativeTemperature, out{ii}.dModelFdT(...
| |
| − | out{ii}.maxModelDerivativeIndex), 'linewidth',2,'marker','x','markersize',18,'color',...
| |
| − | out{ii}.modelPlotColor);
| |
| − | end
| |
| − |
| |
| − | % Add the legends
| |
| − | figure(1)
| |
| − | legend(legendText, 'location', 'southwest');
| |
| − | hold off;
| |
| − | figure(2)
| |
| − | legend(legendText, 'location', 'southwest');
| |
| − | hold off;
| |
| − |
| |
| − | </pre>
| |
| − |
| |
| − | ==Report Requirements==
| |
| − | Report requirements will be posted separately.
| |
| − |
| |
| − | ==References==
| |
| − |
| |
| − | <references />
| |
| − |
| |
| − | ==Datasheets==
| |
| − | #[http://sine.ni.com/nips/cds/view/p/lang/en/nid/207096 National Instruments data acquisition card user manual]
| |
| − | #[http://www.sigmaaldrich.com/sigma-aldrich/datasheet/s9430dat.pdf SYBR Green I datasheet]
| |
| − | #[http://www.national.com/ds/LF/LF411.pdf Op-amp datasheet]
| |
| − | #[http://www.clare.com/home/pdfs.nsf/www/CPC1709.pdf/$file/CPC1709.pdf Relay datasheet]
| |
| − | #[http://www.learn-c.com/tip120.pdf Darlington transistor
| |
| | | | |
| | {{Template:20.309 bottom}} | | {{Template:20.309 bottom}} |