Difference between revisions of "Lab Manual: Measuring DNA Melting Curves"
| Line 1: | Line 1: | ||
| + | [[Category:Lab Manuals]] | ||
| + | [[Category:20.309]] | ||
| + | [[Category:DNA Melting Lab]] | ||
| + | {{Template:20.309}} | ||
| + | |||
| + | {| | ||
| + | |- valign="top" | ||
| + | | __TOC__ | ||
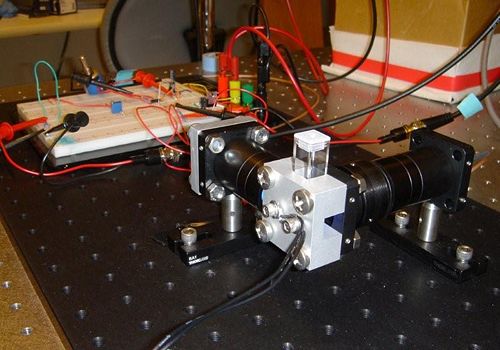
| + | | [[Image:DNA Melting Apparatus Picture.jpg|thumb|center|top|500px|DNA Melting Apparatus]] | ||
| + | |} | ||
| + | |||
| + | ==Introduction== | ||
| + | |||
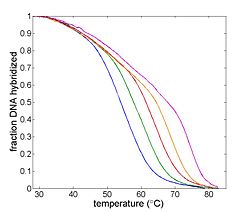
| + | [[Image:Example Melting Curve.jpg|thumb|right|250px|Example DNA melting curves showing the effect of varying ionic strength. The data has been filtered to reduce noise.]] | ||
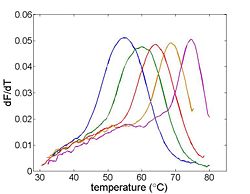
| + | [[Image:Example Melting Curve Derivative.jpg|thumb|right|250px|Differentiating the melting curve simplifies finding T<sub>m</sub>.]] | ||
| + | In this lab, you will build an instrument to measure the thermodynamic properties of DNA samples to determine the effect of sequence length, ionic strength and complementarity. A common application of this technique exploits the length dependence of DNA melting temperatures to examine PCR products in order to determine whether a desired sequence was successfully amplified. | ||
| + | |||
| + | The measurement technique utilizes a fluorescent dye that binds preferentially to double stranded DNA (dsDNA). This characteristic of the dye allows the relative concentration of dsDNA to be determined by measuring the intensity of fluorescent light given off by an excited sample. | ||
| + | |||
| + | The DNA melting apparatus you will construct consists of five major subsystems: excitation, fluorescence measurement, heating, temperature sensing, and data acquisition. Major components of these subsystems include an LED, a photodiode and amplifier, a thermoelectric cooler (TEC), a resistance temperature detector (RTD), and a PC data acquisition system. | ||
| + | |||
| + | The goal of your time in the lab will be to measure fluorescence intensity versus temperature for each of the samples over a range of about 90°C to room temperature. This will provide a basis for estimating the melting temperature, T<sub>m</sub> of each sample. (T<sub>m</sub> is defined as the temperature where half of the DNA in the sample remains hybridized.) | ||
| + | |||
| + | You will measure samples of both known and unknown composition. THe samples may vary in length, complementarity (complete match, single mismatch, or complete mismatch), or salt concentration. You will compare the data you gather to theoretical models and you will attempt to identify unknown samples. | ||
| + | |||
| + | ===Suggested readings and references=== | ||
| + | |||
| + | Zipper H, Brunner H, Bernhagen J, Vitzthum F. Investigations on DNA intercalation and surface binding by SYBR Green I, its structure determination and methodological implications. ''Nucleic Acids Res.'' 2004;'''32''':e103–10.<ref name="SYBR Green Paper">[http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=484200 Zipper H, Brunner H, Bernhagen J, Vitzthum F. Investigations on DNA intercalation and surface binding by SYBR Green I, its structure determination and methodological implications. ''Nucleic Acids Res.'' 2004;'''32''':e103–10.]</ref> | ||
| + | |||
| + | A more complete [[Resource list:DNA melting and PCR|DNA melting and PCR resource list]] is available on this wiki. Please improve the page by adding relevant, high-quality sources. | ||
| + | |||
| + | ===Roadmap === | ||
| + | |||
| + | #Build an optical system containing the LED, heating block, sample, photodiode, filters, and lenses. | ||
| + | #Hook up a three terminal voltage regulator to create an electrical power supply for the LED. | ||
| + | #Build, test, and calibrate the temperature-sensing circuit. | ||
| + | #Build an amplification/offset circuit for the DNA fluorescence signal. | ||
| + | #Troubleshoot and optimize your system. | ||
| + | #Heat a samples of DNA with SYBR Green dye and record DNA melting curves both as the samples heat up, and as they cool. | ||
| + | #Analyze the data: For your known samples, compare your observations to theoretical models. Identify your unknown samples. | ||
| + | |||
| + | |||
| + | ===Objectives and learning goals=== | ||
| + | |||
| + | *Build an optical system for exciting the sample with blue light and gathering the fluorescence output on the photodiode. | ||
| + | *Measure light intensity with a photodiode. | ||
| + | *Build a heating system to reliably heat and cool your sample. | ||
| + | *Measure temperature with an RTD and an appropriate transfer function. | ||
| + | *Implement a high gain transimpedance amplifier. | ||
| + | *Use a lock-in amplifier to reduce noise. | ||
| + | *Record dsDNA concentration versus temperature curves for several samples. | ||
| + | *Analyze the data to find the dsDNA fraction as a function of temperature. | ||
| + | *Estimate T<sub>m</sub> from your data. | ||
| + | *Compare the measured curves with theoretical models. | ||
| + | *Identify unknown DNA samples. | ||
| + | |||
| + | ==Parts of the lab manual== | ||
| + | |||
*[[DNA Melting Part 1: Measuring Temperature and Fluorescence]] | *[[DNA Melting Part 1: Measuring Temperature and Fluorescence]] | ||
*[[DNA Melting Part 2: Lock-in Amplifier and Temperature Control]] | *[[DNA Melting Part 2: Lock-in Amplifier and Temperature Control]] | ||
| Line 4: | Line 62: | ||
*[[Matlab:DNA Melting Data Analysis]] | *[[Matlab:DNA Melting Data Analysis]] | ||
*[[DNA Melting Report Requirements]] | *[[DNA Melting Report Requirements]] | ||
| + | |||
| + | {{Template:20.309 bottom}} | ||
Revision as of 18:29, 28 August 2011
Introduction
In this lab, you will build an instrument to measure the thermodynamic properties of DNA samples to determine the effect of sequence length, ionic strength and complementarity. A common application of this technique exploits the length dependence of DNA melting temperatures to examine PCR products in order to determine whether a desired sequence was successfully amplified.
The measurement technique utilizes a fluorescent dye that binds preferentially to double stranded DNA (dsDNA). This characteristic of the dye allows the relative concentration of dsDNA to be determined by measuring the intensity of fluorescent light given off by an excited sample.
The DNA melting apparatus you will construct consists of five major subsystems: excitation, fluorescence measurement, heating, temperature sensing, and data acquisition. Major components of these subsystems include an LED, a photodiode and amplifier, a thermoelectric cooler (TEC), a resistance temperature detector (RTD), and a PC data acquisition system.
The goal of your time in the lab will be to measure fluorescence intensity versus temperature for each of the samples over a range of about 90°C to room temperature. This will provide a basis for estimating the melting temperature, Tm of each sample. (Tm is defined as the temperature where half of the DNA in the sample remains hybridized.)
You will measure samples of both known and unknown composition. THe samples may vary in length, complementarity (complete match, single mismatch, or complete mismatch), or salt concentration. You will compare the data you gather to theoretical models and you will attempt to identify unknown samples.
Suggested readings and references
Zipper H, Brunner H, Bernhagen J, Vitzthum F. Investigations on DNA intercalation and surface binding by SYBR Green I, its structure determination and methodological implications. Nucleic Acids Res. 2004;32:e103–10.[1]
A more complete DNA melting and PCR resource list is available on this wiki. Please improve the page by adding relevant, high-quality sources.
Roadmap
- Build an optical system containing the LED, heating block, sample, photodiode, filters, and lenses.
- Hook up a three terminal voltage regulator to create an electrical power supply for the LED.
- Build, test, and calibrate the temperature-sensing circuit.
- Build an amplification/offset circuit for the DNA fluorescence signal.
- Troubleshoot and optimize your system.
- Heat a samples of DNA with SYBR Green dye and record DNA melting curves both as the samples heat up, and as they cool.
- Analyze the data: For your known samples, compare your observations to theoretical models. Identify your unknown samples.
Objectives and learning goals
- Build an optical system for exciting the sample with blue light and gathering the fluorescence output on the photodiode.
- Measure light intensity with a photodiode.
- Build a heating system to reliably heat and cool your sample.
- Measure temperature with an RTD and an appropriate transfer function.
- Implement a high gain transimpedance amplifier.
- Use a lock-in amplifier to reduce noise.
- Record dsDNA concentration versus temperature curves for several samples.
- Analyze the data to find the dsDNA fraction as a function of temperature.
- Estimate Tm from your data.
- Compare the measured curves with theoretical models.
- Identify unknown DNA samples.
Parts of the lab manual
Cite error: <ref> tags exist, but no <references/> tag was found