Difference between revisions of "Assignment 9 Overview"
Juliesutton (Talk | contribs) (→Photobleaching) |
Juliesutton (Talk | contribs) |
||
| Line 23: | Line 23: | ||
The goal for Assignment 9 is to write a model for ''V<sub>f,measured</sub>'' that takes these effects into account and use nonlinear regression to estimate the parameters of this function. The model proposed here adheres to Dr. [http://en.wikiquote.org/wiki/George_E._P._Box George E. P. Box's excellent advice] on modeling, in that it is both wrong and useful. Some of the assumptions are more dubious than others. The following reading will guide you through the concepts behind the model which will be useful when you sit down to write your code. | The goal for Assignment 9 is to write a model for ''V<sub>f,measured</sub>'' that takes these effects into account and use nonlinear regression to estimate the parameters of this function. The model proposed here adheres to Dr. [http://en.wikiquote.org/wiki/George_E._P._Box George E. P. Box's excellent advice] on modeling, in that it is both wrong and useful. Some of the assumptions are more dubious than others. The following reading will guide you through the concepts behind the model which will be useful when you sit down to write your code. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
==Finding double stranded DNA fraction from raw data== | ==Finding double stranded DNA fraction from raw data== | ||
Revision as of 17:31, 1 November 2017
Introduction
In Assignment 8, you made some improvements to your DNA melting instrument, and (hopefully) collected some spectacular data. This assignment will focus on extracting useful information from the data in order to make some quantitative conclusions.
The fluorescence voltage, Vf,measured(t), that you measured in lab depends not only on the parameters of interest, ΔH°, and ΔS°, but also on:
- the double stranded DNA concentration Cds(t) (which we know from the outset)
- the dynamics of the temperature cycling system
- thermal quenching of the fluorophore
- photobleaching
- responsivity and offset of the instrument
- binding kinetics of the dye
The goal for Assignment 9 is to write a model for Vf,measured that takes these effects into account and use nonlinear regression to estimate the parameters of this function. The model proposed here adheres to Dr. George E. P. Box's excellent advice on modeling, in that it is both wrong and useful. Some of the assumptions are more dubious than others. The following reading will guide you through the concepts behind the model which will be useful when you sit down to write your code.
Finding double stranded DNA fraction from raw data
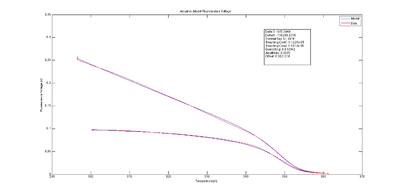
The inverse function of the melting model with respect to Vf,measured(t) is helpful to visualize discrepancies between the model and experimental data caused by random noise in Vf,measured and systematic error in the model Vf,model. The function,
- $ C_{ds,inverse-model}(V_{f,measured}(t)) = \frac{V_{f,measured}(t) - K_{offset}} {K_{gain} S(t) Q(t)} $,
is itself a model. This model estimates the concentration of double stranded DNA based on the observations $ V_{f,measured}(t) $ and the models for bleaching and quenching.
The estimated melting curve may be directly compared with simulations, measurements or other predictions of the true melting curve. The plot at right shows an example of Cds,inverse-model(t) versus Tsample(t). The estimated melting curve is shifted to the right compared to the simulated melting curve, possibly due to systematic error in the sample temperature model. The estimated melting curve also serves as a comparison to the thermodynamic model developed in DNA Melting Thermodynamics, or to any other independent measurement or model of the melting curve, i.e., the concentration of dsDNA vs sample temperature.
Assignment details
In this assignment you will write the code to analyze your DNA melting data in two parts:
- In Part 1, you will define the functions for each phenomenon and combine them into a single fit function;
- In Part 2, you will create some simulated data to verify your code and test your model.
| |
Turn in all of your work (comprehensive list below) on Stellar in a single PDF file named <lastname><firstname>Assignment9.pdf. |
Turn in:
- Assignment 9 Overview
- Part 1: model function
- Part 2: test your code with simulated data
- Part 3: fitting your data
Back to 20.309 Main Page