Difference between revisions of "Assignment 3, Part 1: visualizing actin with fluorescence contrast"
m (Steven Wasserman moved page Assignment 3: visualizing actin using fluorescence contrast to Assignment 3: visualizing actin with fluorescence contrast) |
MAXINE JONAS (Talk | contribs) m (MAXINE JONAS moved page Assignment 3: visualizing actin with fluorescence contrast to Assignment 3, Part 1: visualizing actin with fluorescence contrast) |
(No difference)
| |
Revision as of 19:56, 14 August 2017
|
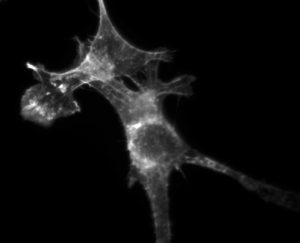
3T3 cells. Actin stained with Alexa Fluor 568 Phalloidin. |
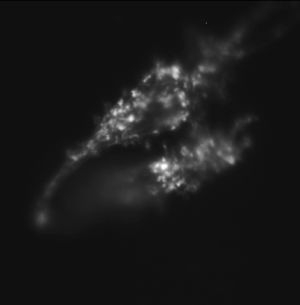
Same cell type incubated with cytochalasin D, an inhibitor of actin polymerization, prior to fixing and stained. |
|---|
Fluorescent staining
In this part of the lab, you will use fluorescence microscopy to investigate the effect of a toxin called cytochalasin D on cultured epithelial cells. Cytochalasin D inhibits the of polymerization of [actin]. Actin is an important component of the cytoskeleton. Exposure to cytochalasin D causes marked changes in the shape and mechanical properties of cells. In this part of the lab, you will make and compare images of healthy (control) cells and identical cells that have been exposed to cytochalasin D. We will investigate some of the mechanical changes in the next part of the lab.
The actin structures we wish to examine is are normally visible under a microscope. There are (at least) three feasible approaches to creating contrast — all involving fluorescence:
- use genetic manipulations to modify the actin protein so that it fluoresces,
- use an antibody conjugated to a fluorescent dye, or
- use a (non-antibody) compound conjugated to a fluorescent dye.
Each of the approaches has advantages and drawbacks. We will use the third approach. The mushroom toxin phalloidin binds to polymerized filamentous actin (F-actin) much more tightly than to actin monomers (G-actin), and stabilizes actin filaments by preventing their depolymerization. You will take advantage of this strong affinity between phalloidin and F-actin to image the cell's actin cytoskeleton. Phalloidin conjugated with the fluorescent dye Alexa Fluor 568 will be introduced in 3T3 mouse fibroblast cells.
By comparing Alexa Fluor phalloidin-stained untreated cells and CytoD-treated cells, you will observe and quantify the effect CytoD has on the actin stress fiber network. Cytochalasin D is also a mycotoxin, but contrary to phalloidin, it poisons the cell by strongly inhibiting actin polymerization.
|
|
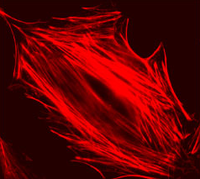
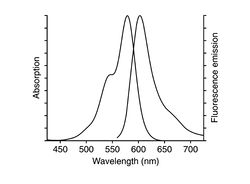
| Example of a NIH 3T3 fibroblast cell labeled with rhodamine phalloidin | Excitation and emission spectra for Alexa Fluor 568 |
| |
Wear gloves when you are handling biological samples. |
Procedures for fixing and labeling cells
You will be given two plates of cells:
- one dish will be left untreated, and then fixed and labeled with phalloidin;
- the second dish will be first treated with CytoD, and then fixed and labeled with phalloidin.
A key technique to keep in mind when working with live cells - to avoid shocking them with "cold" at 20°C - is to be sure that any solutions you add are pre-warmed to 37°C. We will keep a warm-water bath running for this purpose, in which we will keep the various media.
You are provided with 3T3 cells, which were prepared as follows: Cells were cultured at 37°C in 5% CO$ _2 $ in standard T75 flasks in a medium referred to as DMEM++. This consists of Dulbecco's Modified Eagle Medium (DMEM - Invitrogen) supplemented with 10% fetal bovine serum (FBS - Invitrogen), 1% of the antibiotic penicillin-streptomycin (Invitrogen), 1% non-essential amino acids, and 1% glutamine. The day prior to the fluorescence imaging, cells were plated on 35 mm glass-bottom cell culture dishes (MatTek, equipped with coverslip suited for optical microscopy studies).
Please read through the protocol for the two dishes below. Think about what you need to do so you can handle the dishes in parallel and save yourself some time.
Below is the protocol to stain 3T3 cells with Alexa Fluor 568 phalloidin:
- It is optimal for your cells to be ~60% confluent. If cells are too crowded, they will not stretch properly and show their beautiful actin filaments. Thus, you'll want to image cells that are stretched out and not overlapping much. Note also that these cells remain alive until the addition of formaldehyde, therefore requiring that any buffer/media added be pre-warmed.
- Pre-warm 3.7% formaldehyde solution and phosphate buffered saline at pH 7.4 (PBS) in a 37°C water bath if available. Keep the formaldehyde wrapped in foil to protect from light.
- Retrieve an aliquot of cytochalasin D from the freezer and warm to 37°C.
Dish 1--CytoD Treated Cells
- Prepare only ONE dish to be treated with cytochalasin D (CytoD). The other dish will be an untreated control.
- Remove the medium with a pipette and wash ONE dish 2X with 2 mL of pre-warmed PBS. Pipet into the dish gently to avoid washing away cells.
- Add 1 mL of the pre-warmed 10 μM CytoD solution to the same cell culture dish and incubate at 37°C for 20 minutes (and not a minute longer!). Afterwards, wash 2X with PBS.
Dish 1 (CytoD) & Dish 2 (Untreated) Stained in Parallel
- Remove the medium from the untreated dish (Dish 2) with a pipette and wash 2X with 2 mL of pre-warmed PBS. Pipet gently to avoid washing away cells. This assumes you've already washed your Cyto D plate (Dish 1).
- Carefully pipet 400 μL of 3.7% formaldehyde solution onto the cells in the central glass region of each dish and incubate for 10 minutes at room temperature. This "fixes" the cells, i.e. cross-links the intracellular proteins and freezes the cell structure.
- Wash each dish 3X with 1.5 mL PBS (note that this PBS solution no longer needs to be pre-warmed as the cells are dead).
- Extract each dish with 1.5 mL 0.1% Triton X-100 (a detergent) for 3-5 minutes. (Extraction refers to the partially dissolution of the plasma membrane of the cell.)
- Wash the cells 2X with 1.5 mL PBS.
- Incubate the fixed cells with 1.5 mL 1% BSA in PBS for 20 minutes. (BSA blocks the nonspecific binding sites.)
- Wash each dish 2X with PBS.
- Add 200 μL of Alexa Fluor 568 phalloidin solution (pre-mixed in methanol and PBS) to each dish. Carefully pipet this just onto the center of the dish, cover with aluminum foil, and incubate for 45 minutes at room temperature.
- Wash 3X with PBS.
- You can now store the sample at +4°C (regular refrigerator) in PBS for a few days, wrapped in parafilm and foil.
Fluorescence imaging
In this part of the lab, you will make images of fluorescent microspheres plus your dishes of cells, and correct them for nonuniform illumination. In order to do the correction, you will need a reference image and dark images in addition to the image of the sample. See this page for more information about flat-field correction.
- Reference Images: Take the reference image as close as possible in time to the sample images. Don't make any adjustments to your microscope between capturing the reference image and the sample image. For example, every time you move a mirror or re-align the laser, you will change the illumination profile and you must take a new reference. Adjusting the camera exposure and gain between recording the reference and sample images is okay.
- Dark images: Each time you record an image (reference or sample), make sure to take a corresponding dark image using the EXACT SAME camera settings (i.e. use the exact same Exposure Time and Gain settings you had chosen for your reference/sample image). This is the only valid way to subtract the correct dark value from your reference/sample image. Use 12-bit imaging mode to get the best results.
- Saving: Remember to save your images in a format that preserves all 12 bits. We recommend using the MATLAB save command to save data in a .mat file so you can reanalyze it later if necessary. You could also save in an image file format with imwrite. Convert the image to 16-bit, unsigned integer format with the correct range before saving. Read this section of Part 1 if you need to refresh your memory.
Take images
- Record a reference image
- Put in an ND filter and take an image of the reference slide with the 40X objective.
- Turn off the laser and record a dark image (without changing any camera settings!)
- Use MATLAB to generate a histogram of to be certain that the image is exposed correctly.
- Remove the ND filter and image the samples:
- 3.26 μm beads and a corresponding dark image
- Fixed and stained untreated cells and a corresponding dark image
- Fixed and stained cells treated with CytoD and a corresponding dark image
- For each of these samples remember to check the exposure, and to take a corresponding dark image
- Visualizing the actin cytoskeleton under your 20.309 microscope will require skills. Since actin filaments and stress fibers are nanometer-scale objects, they are much dimmer than fluorescent beads or the dye solution - care must be taken to get good images of the cytoskeleton. You may need to cover the microscope to reduce room light contamination. You may find it helpful to use your brightfield microscope to find your cells before turning on your laser illumination, so as to minimize photobleaching of the fluorescent dye.
- Adjust the gain (to zero) and exposure of the camera to get the best picture.
Flat field correction
- Perform flat-field correction on the images.
- Divide the image by a normalized version of your reference image minus the dark image (see this page for more detail).
- Include the original, reference, and flat-field corrected images in your lab report.
Report requirements
General guidelines
- Each part of the microscopy lab requires its own submission. Only the final report will include all 3 sections. You may revise any part of your report until the final deadline. Update the apparatus section to reflect any changes you make during the course of the lab.
- On each due date, one member of your group should submit a single PDF file to Stellar in advance of the deadline. The filename should consist of the last names of all group members, CamelCased, in alphabetical order, followed by a hyphen, the name of the assignment, with a .pdf extension. Example: CrickFranklinWatson-MicroscopyPart1.pdf.
- Use the outline below to break your report into sections.
- Keep your lab report short. Use bullet points, tables, and other organizational tools when appropriate. Brevity is not an excuse for imprecise, incomplete, or unclear communication.
- Be selective in what images you include in your report. Where images are required, include one or a few that convey the character of the dataset. Size images appropriately (does it really need to take up half a page?). At the same time, ensure plots images are readable. A good trick for reducing plot size while maintaining clarity is to adjust the font size and line values so the plot remains clear even when it is small. It helps to save images and plots in an uncompressed format; import and resize them in a way that retains the full quality; and create your final PDF file with settings that do not compress plots and pictures.
- An outstanding error discussion is an essential element of a top-notch report.
- Include any computer code you developed in an appendix at the end of your report. Indicate the source of any code used that you obtained from outside sources.
- Refer to this page for more detail on how to write an excellent report, and to this page for an outline of the final microscopy report.
Part 2 report outline
- Microscope documentation
- Include an updated block diagram of your microscope.
- Images
- Include a figure with an images of the 3.26 μm fluorescent microsphere samples, and the stained cell samples with and without Cyto-D.
- For each sample, create 1 figure with 5 panels.
- The panels of the figure should be: A) unprocessed image; B) reference image; C) dark image; D) flat-field corrected image; and E) histogram.
- In the caption, specify the exposure and gain settings. Each image should have a scale bar. State the dimension of the scale bar in the caption.
- For panel E, plot histograms of the unprocessed, dark, reference, and corrected image on the same set of axes. Plot log10( count ) on the vertical axis and intensity on the horizontal axis. Use a line plot instead of a bar chart for the histogram.
- Image profile
- For one reference, dark and cell image set, plot an intensity profile across the same diagonal. You may also use a bead image, along with it's unique reference and dark images. The intensity of your three images should be on the same scale, i.e., 0 to 65,535 or 0 to 1. Place all three profiles on a single set of axes for comparison. (Use the improfile command in MATLAB.)
- Include a figure with an images of the 3.26 μm fluorescent microsphere samples, and the stained cell samples with and without Cyto-D.
- Discussion
- How did your beam expander design affect your images?
- What differences did you observe between the cells with and without CytoD?
Optical microscopy lab
Code examples and simulations
- Converting Gaussian fit to Rayleigh resolution
- MATLAB: Estimating resolution from a PSF slide image
- Matlab: Scalebars
- Calculating MSD and Diffusion Coefficients