20.109(S22):M2D4
Contents
Introduction
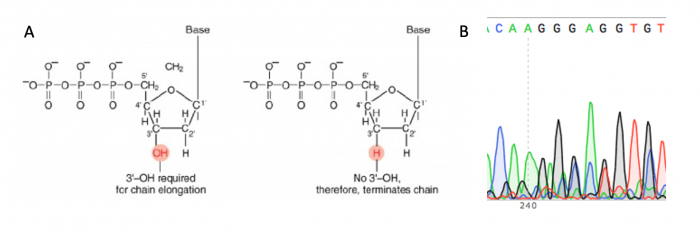
The sgRNA_target sequence that was inserted into the expression plasmid was confirmed using DNA sequencing. The invention of automated sequencing machines has made sequence determination a relatively fast and inexpensive process. The method for sequencing DNA is not new but automation of the process is recent, developed in conjunction with the massive genome sequencing efforts of the 1990s and 2000s. At the heart of sequencing reactions is chemistry worked out by Fred Sanger in the 1970s which uses dideoxynucleotides, or chain-terminating bases. These chain-terminating bases can be added to a growing chain of DNA but cannot be further extended. Performing four reactions, each with a different chain-terminating base, generates fragments of different lengths ending at G, A, T, or C. The fragments, once separated by size, reflect the DNA sequence due to the presence of fluorescent dyes, one color linked to each dideoxy-base. The four colored fragments can be passed through capillaries to a computer that can read the output and trace the color intensities detected.
Protocols
Part 1: Participate in Communication Lab workshop
Our communication instructor, Dr. Prerna Bhargava, will join us today for a discussion on preparing a journal club presentation.
Part 4: Align sgRNA_target sequences to host genome
To ensure that you understand the principles of using sgRNA to target a gene of interest, complete the sgRNA Design Worksheet with your laboratory partner (linked here). It may be helpful to review the work you completed in Part 2 of the previous laboratory session.
In your laboratory notebook, attach the completed sgRNA Design Worksheet.
Next, you will use your knowledge of primer design to align the sgRNA_target sequences that were designed by former 109ers to the targeted genes in the MG1655 genome. Recall that your goal in this module is to optimize the CRISPRi system by building on the data collected by students in previous semesters. The first step in achieving this goal is mining the data that exist! The sgRNA_target sequences that you will assess are included in the table below:
With your laboratory partner, align the sgRNA_target sequences with the targeted genes. Feel free to divide the workload, one partner can align the sequences that target ldhA and the other can align the sequences that target pta-ack.
- Use the KEGG Database to obtain the DNA sequences of the targeted genes (ldhA and pta-ack) in the E. coli K-12 MG1655 strain.
- Enter the name of targeted gene in the Search genes box and click Go.
- Double click on the linked gene name.
- In your laboratory notebook, use the information provided in the KEGG database to answer the following questions:
- What is the full name of the gene (or Definition)?
- In what pathways is the gene involved?
- The amino acid (AA) sequence and nucleotide sequence (NT) for the gene are provided at the bottom of the page.
- Generate a new DNA file in SnapGene that contains the NT sequence of the gene.
- Because sgRNA_target molecules were generated that target the promoter, enter 50 in the +upstream box to get the 50 basepair sequence immediately preceding the start codon.
- Identify the sgRNA_target sequences from the table below in the MG1655 targeted gene.
- For each sgRNA_target sequence, create a feature in the SnapGene file.
- In your laboratory notebook, complete the following:
- Attach the SnapGene file with the sgRNA_target sequences aligned.
- Draw a simplified schematic that shows sgRNA_target sequence alignments. For an example, refer back to Fig. 2C and 2D from the Lei et. al. article.
- Determine which sgRNA_target sequences bind downstream of a PAM sequence and indicate this information on the schematic.
- Speculate on which sgRNA_target sequences might be better at increasing ethanol yield. Include reasons for why!
Reagents list
Next day: Prepare for induction of CRISPRi system