Difference between revisions of "20.109(S22):M2D4"
Noreen Lyell (Talk | contribs) (→Protocols) |
Noreen Lyell (Talk | contribs) (→Introduction) |
||
| Line 6: | Line 6: | ||
==Introduction== | ==Introduction== | ||
| − | The | + | The CRISPRi system involves three genetic components: the pdCas9 plasmid (1 in image below), the psgRNA_target plasmid (2 in image below), and the targeted gene within the host genome (3 in image below). Though the targeted gene is native to the host genome, the plasmids must be transformed into the cell and maintained using antibiotic selection. Thus far in this module, we have discussed the CRISPRi plasmids as individual units, but now we will consider the system as a whole in the context of engineering gene expression. |
| − | [[Image:Fa20 | + | [[Image:Fa20 M3D3 CRISPRi system.png|thumb|center|750px|'''Overview of CRISPRi system.''' The CRISPRi system consists of three genetic components: 1. an expression plasmid that encodes the pdCas9 protein that binds to DNA when complexed with sgRNA, 2. an expression plasmid that encodes the sgRNA that is complementary to the targeted sequence in the host genome, and 3. the targeted sequence in the host genome. Image generated using BioRender.]] |
| + | |||
| + | In the previous laboratory session, you performed the procedure used to generate the psgRNA_target plasmids. Today we will transform the CRISPRi system (pdCasd9 and psgRNA_target) is into ''E. coli''. Once transformed into the bacterial cells, the sgRNA_target and dCas9 are transcribed from the respective expression plasmids. As an overview, the promoter (pJ23119) driving expression of the sgRNA sequence in the gRNA_target plasmid is constitutively active. This means that transcription of the gRNA sequence specific to the target in the host genome is constitutive. Therefore, your sgRNA_target is always present in the MG1655 cells. In a mechanism that we will discuss in the next laboratory session, expression of dCas9 is controlled using an inducer molecule. | ||
<br style="clear:both;"/> | <br style="clear:both;"/> | ||
Revision as of 18:52, 24 January 2022
Contents
Introduction
The CRISPRi system involves three genetic components: the pdCas9 plasmid (1 in image below), the psgRNA_target plasmid (2 in image below), and the targeted gene within the host genome (3 in image below). Though the targeted gene is native to the host genome, the plasmids must be transformed into the cell and maintained using antibiotic selection. Thus far in this module, we have discussed the CRISPRi plasmids as individual units, but now we will consider the system as a whole in the context of engineering gene expression.
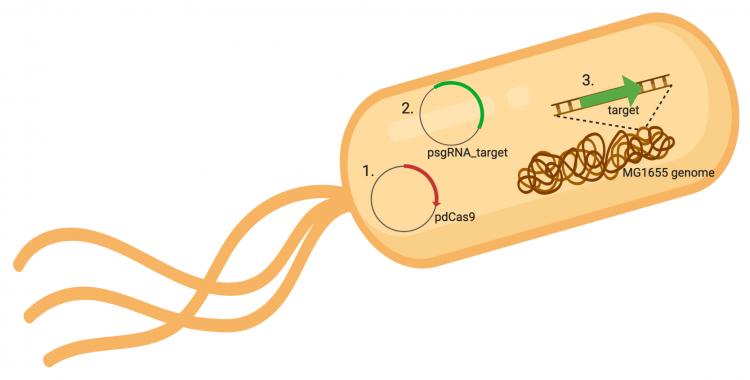
In the previous laboratory session, you performed the procedure used to generate the psgRNA_target plasmids. Today we will transform the CRISPRi system (pdCasd9 and psgRNA_target) is into E. coli. Once transformed into the bacterial cells, the sgRNA_target and dCas9 are transcribed from the respective expression plasmids. As an overview, the promoter (pJ23119) driving expression of the sgRNA sequence in the gRNA_target plasmid is constitutively active. This means that transcription of the gRNA sequence specific to the target in the host genome is constitutive. Therefore, your sgRNA_target is always present in the MG1655 cells. In a mechanism that we will discuss in the next laboratory session, expression of dCas9 is controlled using an inducer molecule.
Protocols
Part 1: Participate in Communication Lab workshop
Our communication instructor, Dr. Prerna Bhargava, will join us today for a discussion on preparing a journal club presentation.
Transform CRISPRi system into MG1655 E. coli cells
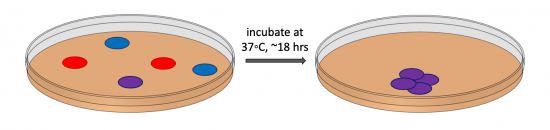
During transformation, a plasmid enters a competent bacterium, then replicates and expresses the encoded genes. In a co-transformation, the goal is to transform each bacterial cell with two plasmids that each encode a different set of genes. Following the co-transformation procedure, a mixed population of cells exists as shown in the figure to the right: some cells only contain the plasmid that carries the resistance cassette for antibiotic A (blue cells), some cells only contain the plasmid that encodes the resistance cassette for antibiotic B (red cells), and some cells contain both plasmids (purple cells). Because the agar plate used for selection contains both antibiotic A and antibiotic B, only bacterial cells that harbor both plasmids survive and reproduce to form a colony.
Most bacteria do not usually exist in a “transformation ready” state, referred to as competence. Instead bacterial cells are incubated with CaCl2 to promote competency by making the cells permeable to plasmid DNA uptake. Competent cells are extremely fragile and should be handled gently, specifically the cells should be kept cold and not vortexed. The transformation procedure is efficient enough for most lab purposes, with efficiencies as high as 109 transformed cells per microgram of DNA, but it is important to realize that even with high efficiency cells only 1 DNA molecule in about 10,000 is successfully transformed.
Reagents list
Next day: Prepare for induction of CRISPRi system