20.109(S16):Cell preparation for DNA repair assays (Day5)
Contents
Introduction
Today you will (finally) use the pMAX_EGFP_MCS reporter construct you prepared and purified during the previous laboratory session with the human glioblastoma cell lines to assess the NHEJ mechanism. Remember that the M059K cells are the wild-type cells, which carry DNA-PKcs, and the M059J cells are DNA-PKcs mutants. This suggests that the M059J cells are unable to perform NHEJ and will, therefore, be the negative control for your experiments. In your experimental setup today you will transfect M059K cells with damaged pMAX_EGFP_MCS and you will also transfect cells with intact, or undamaged, pMAX_EGFP_MCS. The cells transfected with the damaged reporter will be the experimental samples and those transfected with the intact reporter will be used as the positive controls. Let's take a closer look:
- Negative control: The M059J cells are unable to perform NHEJ and therefore cells transfected with damaged reporter will emit background fluorescence and allow you to visualize the level of repair that occurs via another pathway, therefore it is used to show when the NHEJ efficiency is equal to 0%.
- Positive control: The subset of M059K cells that are transfected with intact reporter will show the frequency of fluorescent signal when the NHEJ efficiency is equal to 100%.
- Experimental samples: These are the cells that will give you data!
There are several approaches that researchers have used to introduce DNA into a cell's nucleus. At one extreme there is ballistics. In essence, a small gun is used to shoot the DNA into the cell. This is both technically difficult and inefficient, and so we won't be using this approach! More common approaches are electroporation and lipofection. During electroporation, mammalian cells are mixed with DNA and subjected to a brief pulse of electrical current within a capacitor. The current causes the membranes (which are charged in a polar fashion) to momentarily flip around, making small holes in the cell membrane that the DNA can pass through.
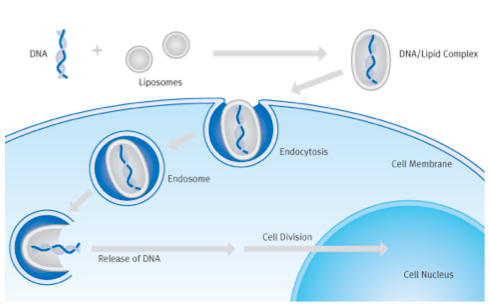
The most popular chemical approach for getting DNA into cells is called lipofection. With this technique, a DNA sample is coated with a special kind of lipid that is able to fuse with mammalian cell membranes. When the coated DNA is mixed with the cells, they engulf it through endocytosis. The DNA stays in the cytoplasm of the cell until the next cell division, at which time the cell’s nuclear membrane dissolves and the DNA has a chance to enter the nucleus.
Tomorrow, 24 hours post-transfection, the teaching faculty will measure the flourescent signal emitted from these cells. Because only cells that carry a repaired plasmid will fluoresce green, you can compare this information to the controls to calculate the NHEJ efficiency in reference to the specific variables you chose/choose. The first variable that you will consider is the type of DNA damage (i.e. the DNA ends created by the restriction enzyme digest). The second variable is the effect of a drug reported to inhibit NHEJ repair in other model systems. The purpose for examining the role of drugs in NHEJ repair efficiency is two-fold: 1) targeting NHEJ inhibitors to cancer cells is important for the treatment of disease, and 2) chemical inhibitors are used widely in biological engineering, so it is useful to think about their strengths and limitations.
Review the following inhibitors, then sign up on the M2D5 Discussion page for the drug that you will use in your assay. Consider collaborating with another team to assess a single drug with two types of DNA damage...or one type of DNA damage with each of the drugs. You will use the drug you choose for two experiments, the flow cytometry-based DNA repair assay, and an irradiation-induced cell-death experiment. The latter is a 'kill curve' that will examine NHEJ inibition with increasing doses of the drug following 4 Gy of irradiation to induce DNA damage. If the drug is effective, the DNA damage will not be repaired efficiently and the cells will not survive.
| Drug | Mechanism of action | Vendor website | Literature reference | Fun fact |
|---|---|---|---|---|
| Loperamide hydrochloride | Unknown (for NHEJ) | Santa Cruz | Goglia et al. | Also known as Imodium |
| DMNB | DNA-PKcs inhibitor | Santa Cruz | Durant et al. | Chemical derivative of vanilla |
Protocols
Part 1: Transfection of cells with plasmid reporters
All manipulations are to be done with sterile technique in the TC facility.
Timing is important for this experiment, so calculate all dilutions and be sure of all manipulations before you begin.
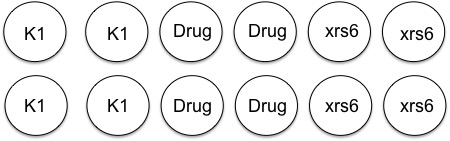
In anticipation of your lipofection experiment, the teaching faculty plated 35,000 K1 and 50,000 xrs6 cells per well in a 24-well dish at ~ 11 am yesterday. Half of the K1 cells then received NHEJ inhibitor at 10am today. A plating schematic for your experiment is shown below. Note carefully which cells are which, including those that have been pre-treated with inhibitor.
Below is a preview of the lipofection workflow. Do NOT begin these steps until you have completed the calculations further below. At each step, be sure to pipet to mix.
- Pre-label all of the eppendorfs that you will need.
- Prepare as much LTX diluted in OptiMEM (hereafter OM) as needed for all 12 transfections, plus 10%.
- The diluted LTX can sit at room temperature during the next two steps.
- Prepare the DNA mixtures – one control (A) and one experimental (B) – diluted in OM, enough for 6 transfections plus 10%, each.
- Add PLUS reagent to the DNA/OM mixtures.
- Distribute the LTX/OM to fresh eppendorf tubes, one for A and one for B. Here include 5% excess.
- For example, if 50 μL were required, you would add 52.5 instead.
- Then add the DNA/OM/PLUS (also 5% excess) to the LTX/OM.
- Incubate for 20 min at room temperature.
- Add 50 μL of the appropriate LTX/DNA/PLUS/OM mixture to each well. Read through the tips below before proceeding.
- Add the LTX/DNA/PLUS/OM drop-by-drop while making a circle with your pipet in the well, and then immediately swirl the plate (circularly) two times.
- Change tips between every addition.
- After distributing to all 12 wells, do a final mixing step for the whole plate, squeaky style: a few each of horizontal and vertical pushes.
The single-well basis reaction for each of the two DNA mixtures is shown below. Scale each one up to enough for 6 wells worth plus excess, carefully following the workflow above – for example, note that not everything is mixed together at once.
| Component | [Stock] ng/μL | Amount per control well (A) | Amount per experimental (B) |
|---|---|---|---|
| LTX (μL) | N/A | 0.5 | 0.5 |
| OM for LTX (μL) | N/A | to total 25 μL per well | to total 25 μL per well |
| intact pMax-GFP (ng) | 100 | 100 | 100 |
| intact pMax-BFP (ng) | 250 | 250 | 0 |
| damaged pMax-BFP-MCS (ng) | unique | 0 | 250 |
| extra cut DNA from Nova (ng) | 50 ng/μL | 0 | to supplement 250 above |
| PLUS (μL) | N/A | 0.5 | 0.5 |
| OM for DNA/PLUS (μL) | N/A | to total 25 μL per well | to total 25 μL per well |
Part 2: Peer Review of Methods Homework
Now that you've had two rounds of feedback and three rounds of drafting practice, you will put your knowledge to work by reviewing and correcting the Methods sections of your peers. First, read the text below as a guide to what your instructors look for while grading Methods sections. Next, read completely through the text of Methods draft that you will offer comments on. Finally, using whatever style is easiest for you, offer specific comments to your fellow 109er about what they have done well and what needs further work. Complete this activity using the "golden rule" -- offer the type of feedback that you hope to also receive. One of the best ways to improve your own skills is to teach something, so take advantage of this opportunity.
Once you have completed your review, please turn in your comments and the Methods section that you reviewed to the front bench. Shannon and Leslie will provide further comments on the Methods section that you reviewed, as well as comments to you as the reviewer to help you with these activities in the future.
(A) Holistic view of Methods section
When the instructors first read your Methods section, we begin by taking a holistic view of its structure. There are three main elements that all good methods sections must contain:
- An appropriate number of sub-sections
- Has the author divided the Methods into logical sub-sections based upon an entire experiment and not based upon completion day in the 20.109 lab?
- Descriptive sub-section titles
- Do the sub-section titles give enough information for the audience to easily pinpoint where the information they need will be found?
- Are the titles specific (i.e. Culture of CHO-K1 and xrs6 cells) or could they belong in any paper that involves cell culture (i.e. Cell culture)?
- Introductory sentences
- Does each sub-section start with an introductory sentence that states the goal of the experiment that was done? (For example, A Western blot was completed to determine Ku80 expression in CHO-K1 and xrs6 cells.)
(B) Specific experimental details
Next, we examine each Methods section to make sure that all of the details that are required to replicate your study are included. For this assignment, the important details are as follows (here we've included them within the sub-sections that would be appropriate for M2D1-D3):
- Cell culture sub-section
- cell types and origin (where did you get them?)
- cell culture media formulation
- culture conditions (temperature, CO2%, and relative humidity)
- Note: later you may choose to include the inhibitor name, concentration, and treatment times in this section.
- Western blot sub-section
- cell seeding density and incubation time
- lysis buffer formulation and lysis conditions (PBS rinse, temperature)
- protein quantification reagent, total protein separated on gel
- SDS-PAGE gel %, TE buffer formulation, electrophoresis conditions
- transfer buffer formulation, transfer conditions, membrane type, blocking buffer
- names of antibodies and dilutions, incubation times
- washing conditions and blot development
- DNA damage sub-section
- name of plasmid, mass digested
- name of enzymes, concentration of enzyme, reaction buffer
- reaction conditions, evaluation by electrophoresis
(C) Quality of writing
Finally, we provide comments on the writing style employed in each Methods section, concentrating on the following:
- Is this a protocol or a formal Methods section?
- Are volumes, vortexing steps, incubation times, etc. that are extraneous (or flexible) included? If so, this is a protocol.
- Is it concise?
- Are "filler words" avoided? Are sentences combined where possible to eliminate extraneous phrases and/or information?
- Is it logical?
- Do the 'steps' follow a logical progression? Again, is the section constructed to explain an experiment as a whole and not simply listed in the order in which it was performed in class?
- Is it written in complete sentences?
- Is the section easy to read or are there words missing/misspelled/misused?
For this activity, please comment on each of the three major evaluation criteria (A-C) listed above. You may do this by following the format that Noreen, Leslie, and Shannon employ -- numbering the paper and then writing an accompanying document. Or, you may simply write comments in a Word doc and provide a copy of that to your Methods section buddy.
Reagent list
- Media as on Day 1
- Lipofectamine® LTX Reagent with PLUS™ Reagent (Life Technologies)
- Opti-MEM reduced serum medium (Life Technologies)
Next day: DNA repair assays
Previous day: Journal Club I