20.109(S14):DNA sequencing (Day5)
Introduction
Last time you reacted your 16S PCR product with specially prepared backbone DNA, and then transformed the reaction product into an engineered cell strain. Eight independent colonies were selected from each of your plates and grown overnight in liquid culture. You will now isolate and sequence DNA from each colony, then pool your results with all other groups studying that particular bird sample to construct a phylogenetic tree representing the bacterial composition in that sample.
We have already discussed the features of the transformation strain needed to synthesize and select for the whole plasmid, namely ccdB gene REVISE. Now let's talk about those features relevant to extracting DNA. As you can gather from the linked manual (PDF), the REVISE cells have endA and recA mutations, both of which make cloning go more smoothly. First, endA1 limits the non-specific destruction of plasmid (and chromosomal) DNA normally carried out by the EndA enzyme, thus maximizing DNA recovery. (The cells also have additional deficiencies in restriction-based nucleases.) Second, recA1 makes the cells incapable of homologous recombination, which could otherwise cause undesirable intermingling between the plasmid and chromosomal DNA.
The procedure for DNA isolation at this scale is commonly termed "mini-prep," which distinguishes it from a “maxi-prep” that involves a larger volume of cells and additional steps of purification. The overall goal of each prep is the same--to separate the plasmid DNA from the chromosomal DNA and cellular debris, allowing the plasmid DNA to be studied further. In the traditional mini-prep protocol, the media is removed from the cells by centrifugation. The cells are resuspended in a solution that contains Tris to buffer the cells and EDTA to bind divalent cations in the lipid bilayer, thereby weakening the cell envelope. A solution of sodium hydroxide and sodium dodecyl sulfate (SDS) is then added. The base denatures the cell’s DNA, both chromosomal and plasmid, while the detergent dissolves the cellular proteins and lipids. The pH of the solution is returned to neutral by adding a mixture of acetic acid and potassium acetate. At neutral pH the SDS precipitates from solution, carrying with it the dissolved proteins and lipids. In addition, the DNA strands renature at neutral pH. The chromosomal DNA, which is much longer than the plasmid DNA, renatures as a tangle that gets trapped in the SDS precipitate. The plasmid DNA renatures normally and stays in solution, effectively separating plasmid DNA from the chromosomal DNA and the proteins and lipids of the cell.
Normally in 20.109 we do an in-house mini-prep procedure according to the steps above followed by ethanol precipitation. However, because you are isolating DNA from so many colonies, today we will use a commercially available kit so that the work can go more quickly. The principle is the same as that of our "quick and dirty" (and cheaper!) prep, but is combined with the silica gel column purification you are familiar with from using other Qiagen kits.
After isolation, you will quantify your DNA by spectrophotometry. Nucleic acids (both RNA and DNA) have an absorbance peak at 260 nm. Beer's law may be used to quantify the amount of DNA from this peak: Abs = ε l c, where Abs is the measured absorbance, l is the path length (1 cm for most specs), c is concentration, and ε is the extinction coefficient. For DNA, ε is 0.02 (μg/mL cm)-1, so 1 absorbance unit corresponds to 50 μg/mL of DNA. The absorbance at 280 nm gives some indication of DNA purity, as proteins have their absorbance peaks at that value (primarily due to the aromatic peptides tryptophan and tyrosine). An Abs260:Abs280 ratio of ~1.8:1 is desired.
Miniprepped DNA will be sent for sequencing off-site. Each clone will be sequenced from two directions, in order to cover the entire 1400 bp region of interest. Together, you'll do about 200 sequencing reactions per lab section!
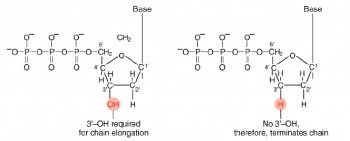
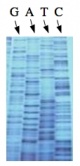
The invention of automated sequencing machines has made sequence determination a relatively fast and inexpensive endeavor. The method for sequencing DNA is not new but automation of the process is recent, developed in conjunction with the massive genome sequencing efforts of the 1990s. At the heart of sequencing reactions is chemistry worked out by Fred Sanger in the 1970s which uses dideoxynucleotides (see schematic above left). These chain-terminating bases can be added to a growing chain of DNA but cannot be further extended. Performing four reactions, each with a different chain-terminating base, generates fragments of different lengths ending at G, A, T, or C. The fragments, once separated by size, reflect the DNA’s sequence. In the “old days” (all of 20 years ago!) radioactive material was incorporated into the elongating DNA fragments so they could be visualized on X-ray film (image above center). More recently, fluorescent dyes have been used instead, with one color linked to each dideoxy-base. The four colored fragments can be passed through capillaries to a computer that can display the color intensities detected (image above right). Your sample was sequenced in this way.
Analysis of sequence data is no small task. “Sequence gazing” can swallow hours of time with little or no results. There are also many web-based programs to decipher patterns. The nucleotide or its translated protein can be examined in this way. Thanks to the genome sequence information that is now available, a new verb, “to BLAST,” has been coined to describe the comparison of your own sequence to sequences from other organisms. BLAST is an acronym for Basic Local Alignment Search Tool, and can be accessed through the National Center for Biotechnology Information (NCBI) home page.
In another week you will finally get to see the results of all your hard work!