20.109(S11):Ligate DNA and transform bacteria (Day4)
Contents
Introduction
FROM S08 and S10 IN REVISION
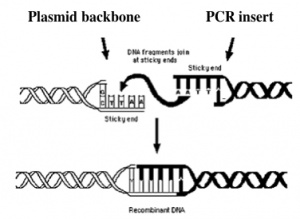
Today you will ligate your linearized and purified plasmid backbone with your unique annealed oligos by mixing the two in the presence of ATP and an enzyme, T4 DNA ligase. During the ligation reactions, hydrogen bonds will form between the overhangs on the fragments, and then the ligase will repair the phosphate backbone, creating a stable circular plasmid (as shown in the figure below).
If all goes well, your ligation reactions will generate your desired construct: the pED-IPTG-INS backbone carrying a modified Plux-λ insert. However, the DNA is present at a low concentration, and we would like to amplify it. Thankfully, we have E. coli bacteria to do this for us quite efficiently! Bacteria can take up foreign DNA in a process called transformation, during which a single plasmid enters a bacterium and, once inside, replicates and expresses the genes it encodes. Most bacteria do not exist in a transformation-ready state, but can be made permeable to foreign DNA by chemical treatment or other means. Cells that are capable of transformation are referred to as competent. Competent cells are extremely fragile and should be handled gently, i.e., kept cold and not vortexed. Bacterial transformation is efficient enough for most lab purposes, resulting in as many as 109 transformed cells per microgram of DNA, but even with highly competent cells only 1 DNA molecule in about 10,000 is successfully transformed. Thus we need a way to identify transformed cells, which is usually accomplished with antiobiotics. For example, the parent vector for the pED plasmids, psB4A3 registry link, also carries a gene that leads to ampicillin-resistance. Consequently, a transformed bacterium will grow on ampicillin-containing agar medium, while untransformed cells will die before they can form a colony (see figure above right).
Protocols
Part 1: Prepare ligation reactions
You will prepare two ligation reactions, one with both backbone and insert, and one with only backbone. The contents will be:
| bkb + insert | bkb only | ||
| Purified backbone | 50-100 ng | 50-100 ng | |
| Annealed oligos | 0 μL | enough for 2:1 molar ratio w/bkb | |
| 10X Ligation Buffer^ | 1 μl | 1 μl | |
| T4 DNA Ligase | 0.5 μl | 0.5 μl | |
| Water | To 10 μl not including volume of enzyme | ||
^New England Biolabs sells 10X Ligation buffer to use with their ligase. It contains ATP so must be kept on ice.
If you cannot meet all of the requirements above given your backbone concentration, it may be okay to use less than 50 ng, or to change the ratio of backbone to insert. Please see the teaching faculty to discuss how to proceed.
- Assemble the reactions in eppendorf tubes but not in the order listed. Please ask if you are unsure what order to assemble the components.
- When the ligation mixes are complete, flick the tubes to mix the contents, quick spin them in the microfuge to bring down any droplets, then incubate the reactions at room temperature for at least 1 hour.
- Even if you will only incubate 1 hour, begin preparing Part 2 (e.g., measure OD600 values and aliquot Z-buffer into tubes). You can finish Part 2 during a later incubation or after you have finished the transformation.
- Alternatively, you can continue the ligation reaction for more than 1 hour until you have finished Part 2. Just keep in mind that there is a later half-hour incubation in Part 3 as well.
- Finally, you and your partner can split up some of the work if that helps, especially since you have all done β-gal assays before.
- Place the reactions on the 65 °C heat block for 10 minutes to heat inactivate the enzyme.
Part 2: Transfer function for IPTG → lacZ
- Follow the protocol from Day 2 for performing β-gal assays.
- Remember to order your samples from least to most expected activity, especially if one person is working without a partner during the ONPG addition step.
Make chart for how they should fill plates
Part 3: Transform bacteria with ligated DNA
You will perform 4 bacterial transformations, one for each of the ligation mixtures, one negative control, and one transformation with 5 ng of plasmid DNA to assess transformation frequency.
- Prewarm and dry 4 LB+AMP plates by placing them in the 37°C incubator, media side up with the lids ajar.
- Get an aliquot of competent cells from one of the teaching faculty. Keep these cells on ice at all times. There should be at least 200 μl of cells in each tube. Aliquot 50 μl of cells into 4 clean eppendorf tubes.
- Add DNA to each tube of cells as indicated below.
- Ligation reactions, 5 μL of appropriate mixture to each
- Positive control, 1 μL of pED-IPTG-INS
- Negative control, none
- Flick to mix the contents and leave the tubes on ice for at least 5 minutes.
- Heat shock the cells at 42°C for 90 seconds exactly and put on ice for two minutes. Use your timer.
- Move the samples to a rack on your bench then use your P1000 to add 0.5 ml of LB media to each eppendorf tube. Invert each tube to mix.
- Place in the 37 °C incubator for 30 minutes.
- Plate 250 μL of each transformation mix on LB+AMP plates. After dipping the glass spreader in the ethanol jar, you should pass it through the flame of the alcohol burner just long enough to ignite the ethanol. After letting the ethanol burn off, the spreader may still be very hot, and it is advisable to tap it gently on a portion of the agar plate without cells in order to equilibrate it with the agar (if it sizzles, it's way too hot). Once the plates are ready, wrap them together with one piece of colored tape and incubate them in the 37°C incubator overnight. One of the teaching faculty will remove them from the incubator and set up liquid cultures for you to use next time.
For next time
plasmid/RE hw from last time (revise?)
some part of report, probably: schematic of design choice, transfer function figure; also introduction or wait until M1 revision?
mid-semester eval (optional, anonymous)
Reagent list
- LB (Luria-Bertani broth)
- 1% Tryptone
- 0.5% Yeast Extract
- 1% NaCl
- autoclaved for sterility
- Ampicillin: 100 mg/mL, aqueous, sterile-filtered
- LB+AMP plates
- LB with 2% agar and 100 μg/ml Ampicillin
- T4 DNA ligase buffer (1X), from NEB
- 50 mM Tris-HCl
- 10 mM MgCl2
- 10 mM DTT
- 1 mM ATP
- 25 μg/ml BSA