Difference between revisions of "20.109(F19):Prepare for cellular thermal shift assay (Day1)"
Noreen Lyell (Talk | contribs) (Created page with "<div style="padding: 10px; width: 820px; border: 5px solid #A4A4A4;"> {{Template:20.109(F19)}} ==Introduction== <font color = red> OVERVIEW OF PREVIOUS STUDENT WORK / EXPERI...") |
Noreen Lyell (Talk | contribs) (→Introduction) |
||
| Line 4: | Line 4: | ||
==Introduction== | ==Introduction== | ||
| − | + | ||
| + | '''Peptidyl-prolyl isomerase (PPIase) assay''' | ||
| + | Today you will test the activity of FKBP12 using a peptidyl-prolyl isomerase (PPIase) assay. PPIases catalyze ''cis''-''trans'' isomerization reactions that are essential to efficient protein folding ''in vivo''. Specifically, these enzymes isomerize peptide bonds that are N-terminal to proline residues in polypeptide chains. Without PPIases, isomerization would be the rate-limiting step in protein folding. | ||
| + | |||
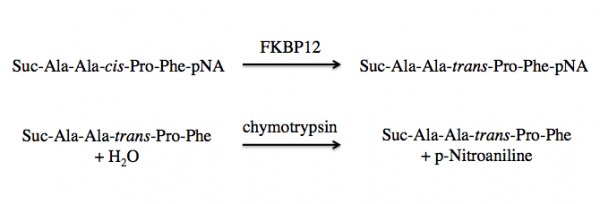
| + | FKBP12 is a class of PPIases and the activity can be measured by quantifying the isomerization and subsequent cleavage of a substrate, suc-AAFP-pNA (also written as Suc-Ala-Ala-Pro-Phe-NA). In this method, FKBP12 catalyzes the isomeration of the ''cis''-Ala-Pro bond to a ''trans''-Ala-Pro bond. Then a second enzyme, chymytrypsin, cleaves the trans form of the peptide. See the reaction schematic below. | ||
| + | |||
| + | [[Image:20.109 Sp18 M2D5 PPiase.png|thumb|600px|center]] | ||
| + | |||
| + | When at equilibrium in solution, approximately 88% of the commercially available suc-AAFP-pNA substrate is in the ''trans'' from, leaving only a small amount of the peptide for isomerization. Because of this, the suc-AAFP-pNA substrate is prepared in trifluoroethanol (TFE) containing lithium chloride, LiCl. The Li+ ions maintain the substrate in 60% ''cis'' form. | ||
| + | |||
| + | Upon cleavage by chymotrypsin, the release of p-nitroaniline results in a yellow color in alkaline conditions, which absorbance can be measured at 405 nm. The rate of ΔA<sub>405 nm</sub> (change in absorbance at 405 nm) in the presence versus absence of FKBP12 is used to calculate the activity. | ||
| + | |||
| + | '''Differential scanning fluorimetry (DSF)''' | ||
| + | Interactions between low molecular weight ligands and proteins have been shown to increase the thermostability of proteins. This means that proteins bound to ligand are able to maintain tertiary structure, or resist denaturation, at higher temperatures than unbound proteins. Today we will use differential scanning fluorimetry (DSF) to examine the potential FKBP12 binders identified in our SMM screen. | ||
| + | |||
| + | DSF is a method used to identify low molecular weight ligands that bind and stabilize a protein of interest. In this assay, protein denaturation is measured via a fluorescent dye that has an affinity for hydrophobic regions. When the protein is folded the hydrophobic pockets are inaccessible to the dye and the fluorescent signal is quenched by water in the solution. As the protein unfolds, the dye interacts with the hydrophobic regions and emits a fluorescent signal that can be detected. | ||
| + | |||
| + | When a protein is bound to a ligand, the stability can be increased such that the temperature at which the protein denatures is increased. In the DSF assay, this is measured as a shift in the T<sub>m</sub>, or melting temperature; which is defined as the temperature at which 50% of the protein is unfolded. This value represents the midpoint of the transition from structured (folded) to denatured (unfolded). | ||
| + | |||
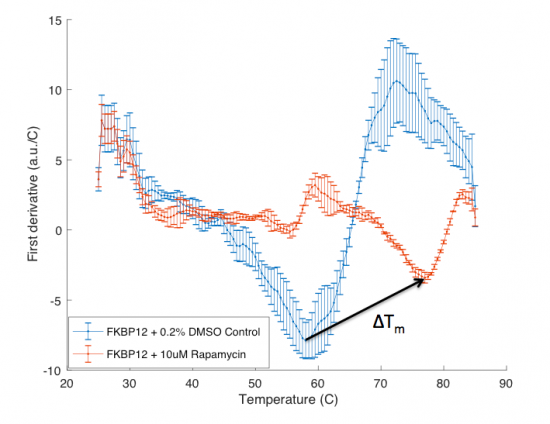
| + | The ΔT<sub>m</sub> is the difference between the T<sub>m</sub> of the unbound protein sample, or protein sample without added ligand, and the bound protein sample, protein sample with added ligand. If the tested ligand binds the protein of interest, the ΔT<sub>m</sub> can be observed as a shift in the plotted DSF data. For example, the data below show results of a pilot experiment completed in preparation for this module. In this graph the T<sub>m</sub> of FKBP12 (blue curve) is ~50 °C. With the addition of rapamycin (red curve) the T<sub>m</sub> is shifted to ~78 °C resulting in a ΔT<sub>m</sub> of ~20 degrees. Data in this plot was obtained by Becky Leifer from the Koehler lab. | ||
| + | [[Image:Sp18 20.109 M1D6 DSF expample data.png|thumb|center|550px]] | ||
==Protocols== | ==Protocols== | ||
Revision as of 18:29, 30 August 2019
Contents
Introduction
Peptidyl-prolyl isomerase (PPIase) assay Today you will test the activity of FKBP12 using a peptidyl-prolyl isomerase (PPIase) assay. PPIases catalyze cis-trans isomerization reactions that are essential to efficient protein folding in vivo. Specifically, these enzymes isomerize peptide bonds that are N-terminal to proline residues in polypeptide chains. Without PPIases, isomerization would be the rate-limiting step in protein folding.
FKBP12 is a class of PPIases and the activity can be measured by quantifying the isomerization and subsequent cleavage of a substrate, suc-AAFP-pNA (also written as Suc-Ala-Ala-Pro-Phe-NA). In this method, FKBP12 catalyzes the isomeration of the cis-Ala-Pro bond to a trans-Ala-Pro bond. Then a second enzyme, chymytrypsin, cleaves the trans form of the peptide. See the reaction schematic below.
When at equilibrium in solution, approximately 88% of the commercially available suc-AAFP-pNA substrate is in the trans from, leaving only a small amount of the peptide for isomerization. Because of this, the suc-AAFP-pNA substrate is prepared in trifluoroethanol (TFE) containing lithium chloride, LiCl. The Li+ ions maintain the substrate in 60% cis form.
Upon cleavage by chymotrypsin, the release of p-nitroaniline results in a yellow color in alkaline conditions, which absorbance can be measured at 405 nm. The rate of ΔA405 nm (change in absorbance at 405 nm) in the presence versus absence of FKBP12 is used to calculate the activity.
Differential scanning fluorimetry (DSF) Interactions between low molecular weight ligands and proteins have been shown to increase the thermostability of proteins. This means that proteins bound to ligand are able to maintain tertiary structure, or resist denaturation, at higher temperatures than unbound proteins. Today we will use differential scanning fluorimetry (DSF) to examine the potential FKBP12 binders identified in our SMM screen.
DSF is a method used to identify low molecular weight ligands that bind and stabilize a protein of interest. In this assay, protein denaturation is measured via a fluorescent dye that has an affinity for hydrophobic regions. When the protein is folded the hydrophobic pockets are inaccessible to the dye and the fluorescent signal is quenched by water in the solution. As the protein unfolds, the dye interacts with the hydrophobic regions and emits a fluorescent signal that can be detected.
When a protein is bound to a ligand, the stability can be increased such that the temperature at which the protein denatures is increased. In the DSF assay, this is measured as a shift in the Tm, or melting temperature; which is defined as the temperature at which 50% of the protein is unfolded. This value represents the midpoint of the transition from structured (folded) to denatured (unfolded).
The ΔTm is the difference between the Tm of the unbound protein sample, or protein sample without added ligand, and the bound protein sample, protein sample with added ligand. If the tested ligand binds the protein of interest, the ΔTm can be observed as a shift in the plotted DSF data. For example, the data below show results of a pilot experiment completed in preparation for this module. In this graph the Tm of FKBP12 (blue curve) is ~50 °C. With the addition of rapamycin (red curve) the Tm is shifted to ~78 °C resulting in a ΔTm of ~20 degrees. Data in this plot was obtained by Becky Leifer from the Koehler lab.
Protocols
Part 1: Communication Lab workshop
Our communication instructors, Dr. Sean Clarke and Dr. Prerna Bhargava, will join us today to provide some tips for your research proposal presentation.
Part 2: Seed cells for CETSA
Part 3: Select ligand to test
Reagents list
Next day: Incubate with ligand and apply heat treatment for protein denaturation