Difference between revisions of "20.109(F16):Module 1"
Noreen Lyell (Talk | contribs) (→Lab links: day by day) |
MAXINE JONAS (Talk | contribs) (→Data) |
||
| (21 intermediate revisions by 3 users not shown) | |||
| Line 8: | Line 8: | ||
'''Instructors:''' [http://be.mit.edu/directory/noreen-lyell Noreen Lyell], [http://be.mit.edu/directory/leslie-mcclain Leslie McClain] and [http://be.mit.edu/directory/maxine-jonas Maxine Jonas] | '''Instructors:''' [http://be.mit.edu/directory/noreen-lyell Noreen Lyell], [http://be.mit.edu/directory/leslie-mcclain Leslie McClain] and [http://be.mit.edu/directory/maxine-jonas Maxine Jonas] | ||
| − | '''TA:''' | + | '''TA:''' Emily Clark <br> |
'''Lab manager:''' Hsinhwa Lee <br> | '''Lab manager:''' Hsinhwa Lee <br> | ||
==Overview== | ==Overview== | ||
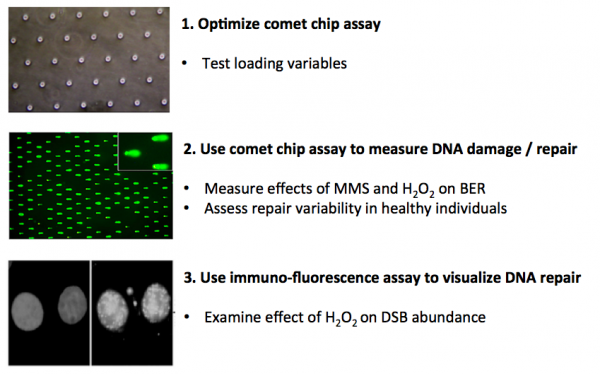
| − | In this module you will measure | + | In this module you will measure genomic instability using two techniques: the CometChip and immunofluorescence. Your first task is to critically think through the development of the CometChip assay and determine which conditions provide the best results for loading mammalian cells into the device. To this end, you will consider variables that affect cell loading into the microwells of the CometChip. The data you collect will be used to determine the conditions for subsequent assays. |
| − | Next, you will use the | + | Next, you will use the CometChip assay to measure DNA damage in response to chemical treatments and to assess repair capacity across different cell lines. Specifically, you will study the effect of oxidative stress in these experiments. Last, you will examine the effect of chemical treatment on the abundance of double-strand breaks using an immunofluorescence approach. |
| Line 22: | Line 22: | ||
==Lab links: day by day== | ==Lab links: day by day== | ||
| − | [[20.109(F16):Prepare comet chips (Day1)| M1D1: Prepare | + | [[20.109(F16):Prepare comet chips (Day1)| M1D1: Prepare microwell array and practice tissue culture]]<br> |
| − | [[20.109(F16):Test comet chip loading variables (Day2)| M1D2: | + | [[20.109(F16):Test comet chip loading variables (Day2)| M1D2: Develop experiment to test loading variables and quantify growth rate]]<br> |
| − | [[20.109(F16):Measure DNA damage with comet chip (Day3)| M1D3: | + | [[20.109(F16):Measure DNA damage with comet chip (Day3)| M1D3: Test role of biochemical factors in genomic stability]]<br> |
| − | [[20.109(F16):Asses DNA repair variability with comet chip (Day4)| M1D4: | + | [[20.109(F16):Asses DNA repair variability with comet chip (Day4)| M1D4: Query inter-individual variability in exposure susceptibility]]<br> |
| − | [[20.109(F16):Seed cells for immuno-flourescence assay (Day5)| M1D5: | + | [[20.109(F16):Seed cells for immuno-flourescence assay (Day5)| M1D5: Develop approach for sub-nuclear visualization of DNA damage]]<br> |
| − | [[20.109(F16):Complete immuno-fluorescence assay (Day6)| M1D6: | + | [[20.109(F16):Complete immuno-fluorescence assay (Day6)| M1D6: Query DNA repair capacity in tumor cells]]<br> |
| − | [[20.109(F16):Data analysis (Day7)| M1D7: | + | [[20.109(F16):Data analysis (Day7)| M1D7: Analysis of sub-nuclear foci]]<br> |
| + | |||
| + | ==Data== | ||
| + | See all M1 student data on the [http://engineerbiology.org/wiki/Talk:20.109(F16):Module_1 Discussion page]. | ||
==Assignments== | ==Assignments== | ||
| − | [[20.109(F16): | + | [[20.109(F16): M1 Data Summary | Data summary]] <br> |
| − | [[20.109(F16): | + | [[20.109(F16): M1 Mini-presentation | Mini-presentation]] <br> |
==References== | ==References== | ||
| − | + | [[Media:CometChip Jove article.pdf| CometChip: A high-throughput 96-well platform for measuring DNA damage in microarrayed human cells.]] ''Journal of Visualized Experiments'' 92: 1-11. | |
| + | A video of the procedure can be found [http://www.jove.com/video/50607/cometchip-high-throughput-96-well-platform-for-measuring-dna-damage here]. | ||
| − | + | [[Media:CometChip-Pages from Ge 2012 Methods in Cell Bio.pdf| CometChip: Single-cell microarray for high-throughput detection of DNA damage.]] ''Methods in Cell Biology'' 112: 247-268. | |
==Notes for teaching faculty== | ==Notes for teaching faculty== | ||
| − | [[20.109(F16): | + | [[20.109(F16): TA notes for M1| F16 notes for M1]] |
| − | [[20.109(F16): | + | [[20.109(F16): TA notes for orientation| F16 notes for orientation day]] |
Latest revision as of 13:43, 23 September 2016
Contents
Module 1
Lecturer: Bevin Engelward
Instructors: Noreen Lyell, Leslie McClain and Maxine Jonas
TA: Emily Clark
Lab manager: Hsinhwa Lee
Overview
In this module you will measure genomic instability using two techniques: the CometChip and immunofluorescence. Your first task is to critically think through the development of the CometChip assay and determine which conditions provide the best results for loading mammalian cells into the device. To this end, you will consider variables that affect cell loading into the microwells of the CometChip. The data you collect will be used to determine the conditions for subsequent assays.
Next, you will use the CometChip assay to measure DNA damage in response to chemical treatments and to assess repair capacity across different cell lines. Specifically, you will study the effect of oxidative stress in these experiments. Last, you will examine the effect of chemical treatment on the abundance of double-strand breaks using an immunofluorescence approach.
Lab links: day by day
M1D1: Prepare microwell array and practice tissue culture
M1D2: Develop experiment to test loading variables and quantify growth rate
M1D3: Test role of biochemical factors in genomic stability
M1D4: Query inter-individual variability in exposure susceptibility
M1D5: Develop approach for sub-nuclear visualization of DNA damage
M1D6: Query DNA repair capacity in tumor cells
M1D7: Analysis of sub-nuclear foci
Data
See all M1 student data on the Discussion page.
Assignments
Data summary
Mini-presentation
References
CometChip: A high-throughput 96-well platform for measuring DNA damage in microarrayed human cells. Journal of Visualized Experiments 92: 1-11. A video of the procedure can be found here.
CometChip: Single-cell microarray for high-throughput detection of DNA damage. Methods in Cell Biology 112: 247-268.