Difference between revisions of "20.109(F16):Induce CRISPRi system (Day7)"
Noreen Lyell (Talk | contribs) (→Introduction) |
Noreen Lyell (Talk | contribs) (→Introduction) |
||
| Line 8: | Line 8: | ||
aTc induction... | aTc induction... | ||
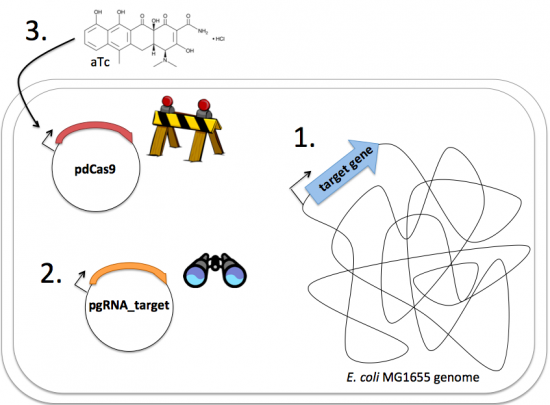
| − | [[Image:Fa16 M2D7 CRISPRi schematic v3.png|thumb|center| | + | [[Image:Fa16 M2D7 CRISPRi schematic v3.png|thumb|center|550px|'''Schematic of CRISPRi system induction.''' Following addition of aTc, the dCas9 protein is produced and recruited by the gRNA target sequence to the target gene. Once recruiting, the dCas9 protein associates with the gRNA/genome complex and impedes transcription by RNAP.]] |
==Protocols== | ==Protocols== | ||
Revision as of 14:46, 12 October 2016
Contents
Introduction
dual plasmid system...
aTc induction...
Protocols
Part 1: Communication Lab workshop
Our communication instructors, Dr. Sean Clarke and Dr. Diana Chien, will join us today for a workshop on organizing and structuring your Research article.
Part 2: Examine pgRNA sequencing results
Your goal today is to analyze the sequencing data for you two potential mutant pgRNA clones - two independent colonies from your amplification reaction - and then decide which colony to proceed with for the CRISPRi manipulation of the E. coli MG1655 fermentation pathway.
Retrieve sequence results from Genewiz
- Use the pgRNA_target Benchling file that you generated on M2D5 to confirm that the gRNA sequence you designed was indeed inserted into the vector.
- Your sequencing data is available from Genewiz.
- Choose the "Login" link and then use "nllyell@mit.edu" and "be20109" to access your results.
- At the bottom right should be a link to download your sequencing results.
- TR section: click on the Tracking Number XXX (Order Date XX/XX/2016).
- WF section: click on the Tracking Number XXX (Order Date XX/XX/2016).
- The quickest way to start working with your data is to follow the "View" link under the Seq File heading. For ambiguous data, you may want to look directly at the Trace File as well.
You should align your sequencing data with a known sequence, in this case the gRNA target sequence you selected, to identify any unintended base changes that may have occurred. There are several web-based programs for aligning sequences and still more programs that can be purchased. The steps for using Benchling are below. Please feel free to use any program with which you are familiar.
Align with Benchling
If both colonies for your mutant have the correct sequence, choose one to use for the co-transformation step. If only one is correct, then this is the one you will use next time. If neither of your plasmids carry the appropriate gRNA target sequence, talk to the teaching faculty.
Part 3: Prepare media for mixed-acid fermentation inoculations
To test the effect of your gRNA on altering the production of either ethanol or lactate, you will incubate the co-transformed E. coli cells both with and without oxygen. Remember from lecture that cells grown anaerobically will produce more fermentation products; however, our goal is to further enhance the production of these products by manipulating the fermentation pathway.
- Obtain a 45 mL aliquot of LB broth from the front laboratory bench.
- Calculate the volume of the chloramphenicol and ampicillin antibiotic stocks that are required to maintain the pdCas9 and pgRNA plasmids, respectively, using the information below:
- For chloramphenicol: the stock concentration is 34 mg/mL and the final or 'working' concentration should be 34 μg/mL.
- For ampicillin: the stock concentration is 100 mg/mL and the final or 'working' concentration should be 100 μg/mL.
- Add the appropriate volume of each antibiotic stock to the LB aliquot and mix thoroughly.
- Acquire 4 glass culture tubes and 4 15 mL conical tubes from the front laboratory bench and label as shown below:
- MG1655 +O2 -aTc (glass tube)
- MG1655 -O2 -aTc (15 mL conical tube)
- MG1655 +O2 +aTc (glass tube)
- MG1655 -O2 +aTc (15 mL conical tube)
- MG1655 +CRISPRi +O2 -aTc (glass tube)
- MG1655 +CRISPRi -O2 -aTc (15 mL conical tube)
- MG1655 +CRISPRi +O2 +aTc (glass tube)
- MG1655 +CRISPRi -O2 +aTc (15 mL conical tube)
- Be sure to include your team information on each tube!
- Transfer 5 mL of the media you prepared in Step #3 to each labelled tube.
On either Monday or Tuesday afternoon, depending on your laboratory section, the teaching faculty will inoculate MG1655 or a co-transformant into your culture tubes and add aTc to the appropriate tubes. All tubes will then be incubated at 37 °C until you return for the next laboratory session.
Reagents
- LB broth
- Luria-Bertani (LB) broth contains 1% tryptone, 0.5% yeast extract, and 1% NaCl
- chloramphenicol antibiotic stock (34 mg/mL)
- ampicillin antibiotic stock (100 mg/mL)
Next day: | Measure fermentation products
Previous day: Journal club II