20.109(S09):Bacterial amplification of DNA (Day3)
Introduction
Assuming all went well, your reaction tubes from last time contain mutagenized DNA that encodes mutant inverse pericam. However, the desired DNA plasmid is likely present at a low concentration, and moreover it is in nicked rather than intact circular form. What we would like to do now is repair and further amplify only the mutagenized product. Thankfully, we have E. coli bacteria to do this for us quite efficiently!
Bacteria can take up foreign DNA in a process called transformation, during which a single plasmid enters a bacterium and, once inside, replicates and expresses the genes it encodes. Most bacteria do not exist in a transformation-ready state, but can be made permeable to foreign DNA by chemical treatment or other means. Cells that are capable of transformation are referred to as competent; competent cells are extremely fragile and should be handled gently, i.e., kept cold and not vortexed.
Bacterial transformation is efficient enough for most lab purposes, resulting in as many as109 transformed cells per microgram of DNA, but even with highly competent cells only 1 DNA molecule in about 10,000 is successfully transformed. Thus we need a way to identify transformed cells, which is usually accomplished with antiobiotics. For example, the plasmid carrying inverse pericam (pRSET) also carries a gene that leads to ampicillin-resistance. Consequently, a transformed bacterium will grow on ampicillin-containing agar medium, while untransformed cells will die before they can form a colony. Given the low concentration and nicked structure of your DNA to begin with, you should perform your transformations today with great care.
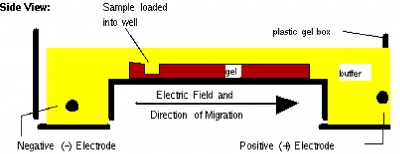
Before setting up transformations, you will test your mutagenized DNA for the presence and approximate concentration of product using gel electrophoresis. This technique separates large molecules by size using an applied electrical field and appropriate sieving matrix. DNA fragments are typically separated in gels composed of agarose, a seaweed-derived polymer. To prepare these gels, molten agarose is poured into a horizontal casting tray containing a comb. Once the agarose has solidified, the comb is removed, leaving wells into which the DNA sample can be loaded. The loaded DNA samples are then pulled through the matrix when a current is applied across it. Specifically, DNA molecules are negatively charged due to their phosphate backbones, and thus travel toward the positive charge at the far end of the gel (see figure).
Although all DNA molecules travel in the same direction during gel electrophoresis, they do so at different rates: larger molecules get entwined in the matrix and retarded, while smaller molecules wind through the matrix more easily and travel further from the well. Ultimately, fragments of similar length accumulate into “bands” in the gel. Bands of DNA are usually visualized by adding the fluorescent dye ethidium bromide to agarose gels. This dye intercalates between the bases of DNA, allowing DNA fragments to be located in the gel under UV light and photographed. The intensity of the band reflects the concentration of molecules that size, although there are upper and lower limits to the sensitivity of dyes. Because of its interaction with DNA, ethidium bromide is a powerful mutagen and will interact with the DNA in your body just as it does with any DNA on a gel. You should always handle all gels and gel equipment with nitrile gloves. Agarose gels with ethidium bromide must be disposed of as hazardous waste.
Today you will run your DpnI-digested mutagenesis reaction mixtures through an agarose gel. In each reaction, the long mutant plasmid DNA should be separated from the short digested fragments of parental DNA and thus can be identified. If you do not see a band at the expected size of the mutant plasmid, you might increase the amount of DNA used during the transformation procedure at the end of lab. In between electrophoresis and transformation, we will (most likely have visit from WAC today)…