20.109(S07): Measuring calcium in vitro
Contents
Introduction
When we learned about M13, we saw how cleverly this virus interacts with its bacterial host, for example using a natural and common bacterial structure to gain access to the cell and then harnessing the cell’s transcription and translation machinery to make more viral particles. What we did not consider was the bacterial response to the viral attack. In fact, bacteria do “notice” viral infection and mount an SOS response within minutes. Stress response genes are up-regulated in an effort to protect the bacterial host from far more severe consequences of phage infection like lysis or destruction of it’s own genome.
In this next module we will turn our attention from the stress-or to the stress-ee, and consider the signals that go on inside cells to help them understand their world. It’s a complex world and so not surprisingly there exist an impressive volume and diversity of cellular messengers. Some relay information between a cell’s outer membrane and its internal processors. Messages in eukaryotic cells also travel between the nucleus and the cytoplasm, plus between and within organelles. Signaling information is transmitted through covalent modifications of the cell’s macromolecules. Common modifications include phosphate-, methyl-, and acetyl- groups, as well as peptide tags to target proteins for particular cellular locations or for degradation. The cellular signal we will consider for the next experimental module is calcium, which is not covalently added to proteins but rather modulates the shape of the proteins that bind it.
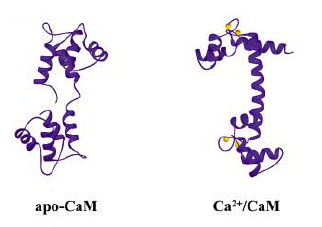
Calcium is the most abundant mineral in our body yet the concentration of Ca2+ within our cells is extremely low (~10^-7M). Calcium pumps and gated channels work hard to establish and keep this steep gradient. Inside the cell, calcium is spatially restricted, stored in some organelles and not others, all poised for release when appropriate. Calcium-handling proteins are a large and widely distributed family of proteins. Many share a characteristic dumbbell shape, with two globular domains connected by a flexible linker region. You'll remember calmodulin, the calcium sensing protein whose structure you examined last time, that binds four Ca2+ ions, two with high affinity and two with low affinity. The high affinity sites can be filled at low calcium concentrations, but when calcium gets released from intracellular stores, the low affinity sites are also filled, inducing a large conformational change in the protein. The change exposes non-polar regions of calmodulin that can bind to non-polar regions of a target molecule. Notice how the backbone of calmodulin extends in the presence of calcium (figure from the review by Vetter and LeClerc) in FEBS 2003.
An interesting example of this is seen with anthrax toxin, which exploits both the cellular distribution of calmodulin as well as its mechanism for calcium sensing. One component of the anthrax toxin, called EF, can bind to calmodulin in the “low calcium” conformation, twisting the linker so it cannot respond to high calcium concentrations, effectively blocking its natural signaling functions. With calmodulin out of the picture, the cells have no way to convert the cAMP formed by EF back into ATP, leading to depletion of the cell’s energy stores. This mechanism seems particularly clever when you remember that calcium-sensing motifs are widespread and abundant but only in animal cells. Thus the Bacillus anthracis bacteria don’t deplete their own energy stores when anthrax toxin is expressed.
Today we will measure calcium concentrations of several solutions. Beyond emphasizing some important laboratory techniques (like standard curves and spectrophotometric analysis of chromogenic reactions), today’s work should underscore the limited sensitivity and range of such techniques. Luckily cells are better at sensing calcium that we are in the lab, as we’ll see when we look at the genetically encoded calcium sensor next week.
Protocols
Part 1: In vitro Ca2+ measurements
We will be using a calcium assay kit sold by "BioAssay Systems" (catalog #DICA-500) that's more commonly applied to diagnose kidney, metabolic and blood disorders. Samples (urine, saliva, serum or in our case CaCl2 solutions) are mixed with a dye that forms a blue color in the presence of free Ca2+. The intensity of the blue color directly reflects the concentration of Ca2+, within the linear detection range of the kit. This range is reported to be 20 uM to 5 mM. You will directly measure the linearity of the assay within this range and beyond. Then you will determine the concentration of free Ca2+ in solutions that are relevant to your tissue culture experiment.
- Make 50 ml of a CaCl2 stock solution (fw 110.99) that is 4X more concentrated than the highest value stated as linear in the BioAssay Systems kit. Use only the good water from the back room of the lab since this will not have free calcium contaminating it. Alternatively, you can try to use the tap water as a point of comparison to others in the lab. It might be interesting to know just how much Ca2+ you can measure this way.
- Dilute this stock in 2X steps, using a 96 well dish or in eppendorf tubes. You should make 100 ul of each dilution. Stop when you are 4X lower than the lowest concentration stated as linear in the BioAssay kit. Be sure to mix each dilution before making the next in the series, and change tips between dilutions.
- Move 5 ul of each dilution to a cuvette, noting which cuvette has which dilution.
- You should also set up a triplicate set cuvettes with 5 ul of a solution relevant to the mouse embryonic stem cells. This could be Growth Media, Trypsin solution, OptiMeM etc. Alternatively, request an unknown from one of the teaching faculty.
- Set up a table in your notebook to record the spectrophotometric data you will collect. The table should include a row for sample descriptions and another for the absorbance values you'll measure at 612nm. You could also include a column to record any observations you have of any sample (color, clarity etc).
- In a 15 ml conical tube, mix 5 ml of BioAssay Reagent A with 5 ml of BioAssay Reagent B.
- Add 500 ul of the assay mixture to each cuvette and mix by inverting several times using a strip of parafilm or a gloved finger to cover the top of the cuvette.
- Incubate at room temperature for at least 3 minutes. Samples are stable for more than an hour.
- Use 500 ul of assay mixture to blank the spectrophotometer (612nm) and than record each sample you have prepared.
- Enter the data you've collected into an Excel spreadsheet. Then:
- prepare a graph with all the standards you measured. What is the equation for the best fit line? What is the R-squared (a measure of linearity). Print out a graph for you lab notebook.
- prepare a graph with only the concentrations of CaCl2 that fall within the stated linear range of the BioAssay Kit. What is the equation for the best fit line? What is the R-squared value? Print out a graph for you lab notebook.
- Using the best fit line you most trust (state which and why you chose that one), determine the concentration of calcium in the cell culture solution you chose. How well do the triplicate measurements agree with one another?
- Enter the requested information onto the discussion page associated with today's lab.
Part 2: Cell culture
You will spend another part of lab today in the cell culture facility, moving MES cells to slides. This protocol is quite similar to the one from last time (so if you need reminding, refer back to the directions from last time). There are two major differences in today's protocol. One is that you will resuspend the cells in media that lacks antibiotics. This is essential to prepare the cells for transfection, but it puts your cells at additional risk for contamination. You'll have to use your very best asceptic technique. Another important difference today is that you will be growing the cells on slides rather than in flasks or dishes. This will allow you to better observe the cells later in this experimental module with the fluorescent microscope.You and your partner should each prepare a slide, though you can share the cells from one flask.
- Begin by preparing the cell culture hood as you did last time.
- Pregelatinize each chamber of a 4-well slide, using 0.5 ml of gelatin dispensed from a 2 ml pipet.
- Retrieve one flask of cells from the incubator.
- Aspirate the media.
- Wash the cells with 5 ml PBS.
- Add 2 ml of trypsin for one minute precisely, aspirate the trypsin, then incubate the "dry" flask in the 37° incubator for ten minutes precisely.
- Triterate the cells with 5 ml "pre-transformation media."
- Dilute the cells 1:20 in pre-transformation media. You will need only 2 ml of dilution per slide. You and your partner can share one dilution mix.
- Aspirate the gelatin from each well of the slides you've prepared.
- Aliquot 0.5 ml per well.
- Place your slide in a petri-dish that can be returned to the incubator until next time. One of the teaching faculty will feed your cells the day before you return to lab.
DONE!
For next time
- Finish filling in the table that is associated with today's lab if you didn't have time to finish in lab.
- Next time you will transfect the cells you plated today. Timing and concentrations are very important for this part of the experiment. Consequently, you should read ahead to the protocol of the next lab.
Reagents list
- QuantiChrom(TM) Calcium Assay Kit (DICA-500) from BioAssay Systems
- Pre-transformation media
- 500 ml DMEM (high glucose)
- 50 ml serum (FBS from Atlanta Biol)
- 5 ml 100X glutamine
- 1 ml BME
- 5 ml non-essential amino acids
- filter then add 50 ul LIF. Store 4° for no more than 1 month.