20.109(S16):Complete Western and prepare damaged DNA (Day3)
Introduction
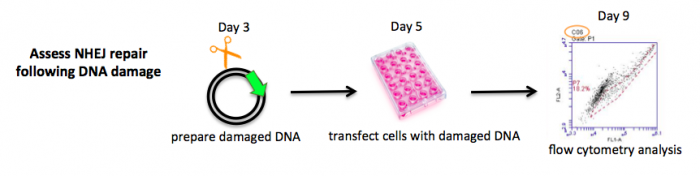
Measuring NHEJ repair
In Module 1 we discussed two uses for restriction enzymes: digesting DNA fragments in preparation for ligation and digesting DNA products to confirm cloning. Here we will use restriction enzymes to generate damaged DNA for your NHEJ assay. Specifically, the damaged DNA will serve as a substrate for NHEJ in the M059K and M059J cells. Because dsb repairs result in a fluorescent signal, you will use a flow cytometer to count the number of cells that successfully repaired the damage caused by the restriction enzyme(s). Moreso than just measuring NHEJ activity in the cell lines, you will also investigate the impact of the type of DNA damage on NHEJ repair.
You will begin today by digesting pMAX-EGFP-MCS using the restriction enzyme(s) you selected previously. Introducing restriction enzyme site-based damage into DNA is just one creative way that scientists and engineers have harnessed the power of nature. In both Module 1 and this current module, you have witnessed molecules (such as ligase) and cells (such as E. coli) stripped from their natural context and applied to solve a problem. Though it now seems mundane, this practice of purifying and re-engineering biology for human ends is pretty amazing if you think about it! Several of the enzymes that will be used in class are “high fidelity” variants, designed for more specific activity than nature requires. Other enzymes have been engineered for faster kinetics.
The way that we are using restriction enzymes in Module 2, while not unprecedented, is somewhat unusual. You already performed the two far more common laboratory practices that utilize restriction enzymes. One is restriction-based cloning of distinct pieces of DNA with compatible ends. Remember, an insert and a vector are ligated together, and then amplified in bacteria. Also, individual DNA clones are miniprepped and tested for correctness. In this second practice, restriction enzymes are just as useful in confirmation or diagnostic digests as they are in creating a clone in the first place. You might be wondering why researchers go through the trouble of designing and performing diagnostic digests, when sequencing is relatively simple and yields more information. Here, the idea of scale becomes important. Sequencing costs $6-8 per reaction, which can add up if you need to examine, say, 10 or more candidates. Agarose gel electrophoresis, by comparison, costs perhaps $1 per candidate. Since both methods require DNA isolation, one is not dramatically more labor intensive than the other. Finally, banding patterns can give a quick readout of many candidate colonies at once compared to the time it takes for individual sequencing analyses (at least if performed manually).
After digesting pMAX-EGFP-MCS to generate damaged ends, you will evaluate the completeness of the digestion on a gel. This step will ensure that the plasmid was digested and also allow you to separate the plasmid from the nonsense insert via gel purification. You already know plenty about agarose electrophoresis, and a fair bit about purifying DNA as well. In gel purification, the agarose containing the DNA of interest is excised from the gel and melted. The DNA is then isolated on a silica (SiO2) column similar to the ones you used in Module 1. Salt concentration and pH effects, along with ethanol precipitation, will alternately allow for binding and eluting the DNA while washing away contaminants.
Measuring protein concentration
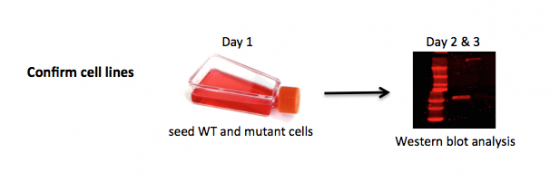
In addition to preparing your damaged DNA for the NHEJ repair assay, you will also complete your Western blot analysis. Your blot was probed with two primary antibodies overnight by the teaching faculty - α-DNA-PK and α-α-tubulin. The α-DNA-PK antibody was raised in a mouse and the α-α-tubulin antibody was raised in a rabbit. Today you will add the two secondary antibodies that will allow for detection.
Traditionally, only one species of antibody could be used on a Western blot because the detection relied on the emission of light that was collected by x-ray film. In the traditional systems the output looks like black bands on a blue or clear background. However, more recent conjugate chemistry has allowed secondary antibodies to be coupled to fluorescent tags. Today we will use infrared (IR) secondary antibodies to detect our α-DNA-PK and α-tubulin antibodies and then scan the Western blots using a specially constructed microscope located in the Lauffenburger lab to determine the level of DNA-PK in our cell lines.
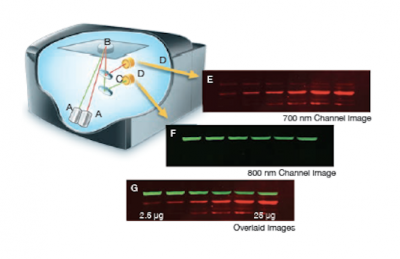
The Licor Odyssey scanner consists of an inverted microscope with two lasers that excite dyes which emit light in the IR range. As depicted in the image on the right, an excitation point is created when beams from the 700 nm and 800 nm lasers (A) are focused on the scanning surface. The microscope objective (B) is focused on the excitation point and collects light from the fluorescing IR dyes. This light is passed through a dichroic mirror (C) that separates the light into two distinct signals that travel through two independent optical paths that are focused on separate silicon photodiodes (D) and detected. In the image, the first channel (E) and second channel (F) are shown separately and merged (G).We will detect our IR-dye conjugated secondary antibodies at wavelengths of 700 and 800 nm. The 700 nm channel will appear red and the 800 nm channel will appear green. Infrared secondary antibodies provide a more flexible detection platform than the traditional Western blot detection methods that rely on colorimetric or chemiluminescent substrates. Unlike the colorimetric or chemiluminescent detection methods, IR dyes do not require a chemical reaction to occur in order for signal to be detected. This means that the output signal increases with time as the colorimetric or chemiluminescent substrate reaction proceeds -- making timing an important variable in traditional Western blot development. We remove that variable from the equation and control when we want to visualize our Western blot simply by controlling the excitation of the dye.
Protocols
Today you get to experience a bit of graduate student life in that you are juggling multiple assays with staggered incubation times. Be sure not to forget about your digesting DNA while completing your Western blot analysis, or about your developing Western blot while gel purifying your damaged DNA!
Part 1: Prepare damaged DNA for NHEJ assay
You will digest pMAX-EGFP-MCS using the enzymes that generate the damaged ends you chose to investigate last time. You will then evaluate the completion of the digestion reaction using gel electrophoresis. Lastly, you will purify the damaged DNA from the agarose for your NHEJ assay.
- Prepare your digestion reaction by combining 7.0 μg of DNA with water, buffer, and enzyme in a labeled eppendorf tube.
- See the M2D3 homework and/or talk to your instructors for more details!
- To avoid pipetting very small volumes, you will either prepare a reaction cocktail that uses no less than 1 μL of the restriction enzyme(s), or you will prepare an intermediate dilution of the enzyme(s).
- Note that enzyme stock concentrations can be found on the NEB product page for that enzyme.
- Remember, the enzyme should be added to the reaction mix last.
- Why? What would happen if you added the enzyme directly to water?
- Flick the eppendorf tube to mix and use the centrifuge to collect the liquid at the bottom of the tube.
- Incubate the digest reaction at 37°C for at least one hour.
- Write down your start time and also set a timer, in case you get distracted.
- While your sample is digesting, you should proceed to the Western blot analysis in Part 2.
Part 2: Complete Western blot protein analysis
Last time, you prepared protein extracts from M059K and M059J cells, separated them using SDS-PAGE, and then transferred them to a nitrocellulose membrane. Following the transfer step, blots were moved to blocking buffer. The next day, the teaching faculty added the α-DNA-PK and α-tubulin primary antibodies and incubated the membranes overnight at 4 °C.
- Obtain your blots from the front bench. Pour the antibody solution into a conical tube, writing the identity of the antibodies and the date on the tube.
- Because the antibody is in excess, the diluted primary solution may be re-used on another blot and is thus worth saving until you see your Western blot.
- Add enough TBS-T to cover your membrane - no need to measure a volume.
- Keep in mind that the washing steps work by dilution, so it is a balance between adding enough to create a sink for the primary antibody, but not so much that you make a huge mess on the shaker!
- TBS-T stands for Tris-buffered saline with 0.1% Tween 20 (a surfactant).
- Shake your container for 5 min at 80 rpm using the room temperature shaker.
- Repeat for a total of 4 washes.
- Just before pouring off the last wash, prepare the secondary antibodies.
- Dilute the secondary antibodies in 10 mL of Blocking Buffer.
- They are light sensitive so find them on the front bench next to the Blocking Buffer and then wrap your tube in aluminum foil.
- For DNA-PKcs (mouse)/ α-tubulin (rabbit) -- use the goat anti-mouse IR800 (GREEN) antibody at 1:10,000 + donkey anti-rabbit IR680 (RED) at 1:10,000.
- After the last wash, add your secondary antibody solution, place on the room temperature shaker, and cover your Western blot container with aluminum foil.
- Shake at 65 rpm for 60 min.
- Pour off the secondary antibody in the sink.
- Wash the membrane by adding TBS-T and shake for 5 min at 80 rpm, using the room temperature shaker.
- Repeat for a total of 4 washes.
- The Odyssey scanner is located in the Lauffenburger laboratory (56-378), one of the teaching faculty will accompany you there in groups to scan your blots.
Part 3: Gel electrophoresis of damaged DNA for NHEJ assay
- When your digest is complete, add 5 μL of 6x NEB loading dye to it, and then pipet 27 μL into a 1% gel according to the scheme in the table below.
- The gels will be run for 30 minutes at 100 V, which should allow for sufficient separation between the plasmid DNA fragment of interest and the nonsense insert.
- To prepare for Part 4, label and weigh an eppendorf tube.
- It is best to write the weight of the empty tube right on the tube.
- After your gel run is finished, the teaching faculty will show you how to safely excise the damaged DNA from your agarose gel.
Four groups will share one gel. We are leaving space between the samples for two reasons:
- We don't want the differently digested DNA bands to bleed into each other, but rather to be well separated.
- We don't want to expose the bands that will be excised later to excess UV. Why?
| Lane | Sample (27 μL) | Lane | Sample (27 μL) |
|---|---|---|---|
| 1 | Group 1 | 6 | BLANK |
| 2 | BLANK | 7 | Group 3 |
| 3 | Group 2 | 8 | BLANK |
| 4 | BLANK | 9 | Group 4 |
| 5 | DNA ladder (load 10 μL) | 10 | BLANK |
Part 4: Gel purification of damaged DNA for NHEJ assay
To purify your DNA from the agarose, you will use a kit from the Qiagen company. As we learned during Module 1, reagents in such commercial kits can have uninformative names and their contents are in part proprietary.
- Estimate the volume of your gel slice by weighing it.
- Add your excised agarose to the eppendorf tube you pre-weighed in Step 3. Then weigh the eppendorf tube with the excised agarose and calculate the difference.
- What does this method assume about the density of agarose and why?
- Add 3 volumes of QG for every 1 volume of agarose.
- The maximum advised volume is 550 μL. If you have a greater volume, continue in one tube for now, but first read step 6 to understand how to proceed later. Feel free to ask the teaching faculty for clarification.
- Incubate in the 50°C water bath for 10 minutes, until the agarose is completely dissolved.
- Every few minutes, you should remove your tube from the 50°C heat and flick or vortex it for a few seconds to help dissolve the agarose.
- Add 1 volume — original gel volume, not current solution volume — of isopropanol to the dissolved sample and pipet well to mix.
- Get one QIAquick column and one collection tube from the front bench.
- Label the spin-column (not the collection tube!) with your team color.
- Carefully pipet the dissolved agarose mixture onto the column. Microfuge for 60 seconds at maximum speed (approx. 16,000 rcf). The maximum capacity of the QIAquick columns is 800 μL! If you have more than 800 μL in your mixture, you will need to repeat this step using the same column.
- Discard the flow-through in a temporary waste conical tube and replace the spin-column in the collection tubes.
- Add 500 μL of QG to the top of the column and spin as before.
- Discard the flow-through.
- Add 750 μL of PE to the top of the column and incubate for 5 min at room temperature.
- Spin for 1 min at max speed.
- Discard the flow-through once more and replace the spin-column in its collection tube.
- Add nothing to the top but spin for 60 seconds to dry the membrane.
- This step completely removes remaining ethanol. Why is this important?
- Meanwhile, trim the cap off of a fresh eppendorf tube, and clearly label the cap with you team color and section.
- Place the labeled spin-column in the de-capped eppendorf tube and add 30 μL of distilled water pH 8.0 to the center of the membrane.
- Allow the column to sit at room temperature for 5 min and then spin as before. The material that collects in the bottom of the eppendorf tube is your purified, digested DNA.
- Alert the teaching faculty when you have completed the DNA purification. You will measure your DNA using a Nanodrop, which reliably evaluates nucleic acid concentrations using a very small sample volume.
- Because your sample is precious and we don't have much to spare, using enough sample to get a good reading on our DU640 spectrophotometer would not be a great idea today!
Reagent list
DNA digestion
- Enzymes from New England Biolabs (NEB).
- Agarose gels: 1% in TAE (see Module 1 for details).
- QIAquick gel extraction kit from Qiagen.
Western blot analysis
Used in steps completed by teaching faculty:
- Odyssey blocking buffer from Licor
- α-DNA-PK primary antibody from BD Transduction Laboratories; used at a 1:1000 dilution of stock.
- α-tubulin primary antibody from Proteintech; used at a 1:5000 dilution of stock.
Used today:
- Donkey anti-Rabbit IR680 Antibody from Licor.
- Donkey anti-Mouse IR800 Antibody from Licor.
- TBS-T
- 50 mM Tris-Cl, to pH 7.5
- 150 mM NaCl
- Tween 20 at a final concentration of 0.1%
- Licor Odyssey System for Western blot detection.
Next day: Journal Club I
Previous day: Begin Western protein analysis and choose system conditions