20.109(S12):Purify RNA and run affinity column (Day4)
Contents
Introduction
Today will be a busy day! Last time you prepared the 6-5 and 8-12 putative aptamers by an in vitro transcription reaction. Now you must purify the RNA before enriching the heme-binding aptamer on an affinity column.
To isolate RNA and change buffers, you will again do a column purification. However, this time the matrix will be a polyacrylamide gel rather than a silica resin. This size exclusion matrix retains nucleic acids shorter than 20 bases. After purification, you will quantify your RNA by spectrophotometry. Nucleic acids (both RNA and DNA) have an absorbance peak at 260 nm. Beer's law may be used to quantify the amount of RNA from this peak: Abs = ε l c, where Abs is the measured absorbance, l is the path length (1 cm for most specs), c is concentration, and ε is the extinction coefficient. For RNA, ε is 0.025 (μg/mL cm)-1, so 1 absorbance unit corresponds to 40 μg/mL of RNA. The absorbance at 280 nm gives some indication of RNA purity, as proteins have their absorbance peaks at that value (primarily due to the aromatic peptides tryptophan and tyrosine). An Abs260:Abs280 ratio of ~2:1 is desired. As a final preparation before column selection, the appropriate amount of RNA must be denatured at 70 °C and re-natured at room temperature in order to ensure that it has the right secondary structure. Otherwise the 8-12 aptamer might not bind to heme.
By now you should have signed up for your experimental conditions on today's Talk page. You and your partner will use the same ratio of the two RNAs, but a different number of column washes. The affinity column you will use consists of agarose beads with bound heme. Thus, when you add your RNA mixture to the column, 8-12 should bind but 6-5 should not. Of course, some 6-5 will non-specifically bind to the column. To remove it you will add aliquots of selection buffer to the column and let the 6-5 wash away. If you wash too vigorously, you may lose some 8-12 as well. One purpose of today's experiment is to hone in on the ideal wash conditions, that is, conditions that retain 8-12 well on the column relative to 6-5. Finally, to collect aptamer that remains bound to the column, you will add a concentrated heme solution. The free heme will compete with the bound heme for aptamer binding, and the aptamers should be readily eluted after a brief incubation step. One additional complication is that nucleic acids bind non-specifically to agarose. Thus, we will use tRNA in our buffer to compete with the aptamers for these non-specific sites. Overall, 8-12 aptamer should be enriched compared to 6-5 by the end of this process.
Once you have collected your samples, you will begin the process of concentrating the collected RNA. A common way to do this is by salt and ethanol precipitation. The ethanol lowers charge-charge screening, allowing the positive ion of the salt to ionically bind and precipitate out the negatively charged nucleic acids. After the column selection, your samples may contain little RNA, and should benefit from a long precipitation step (until our next session). You will also try to limit nucleic acid losses by adding a 'carrier' that co-precipitates with the RNA, namely glycogen. Ultimately, to see how the selected RNA performs in a binding assay you will first have to amplify it.
Protocols
Before starting today, make sure to clean your bench area, pipettes, and centrifuge with RNase away.
Part 1: Prepare spin purification columns and purify RNA
- Take your thawed IVTs from last time, and add 1/10 of the reaction volume of DNase to each reaction.
- For example, for a 100 μL reaction, you would add 10 μL of DNase.
- What is the purpose of adding the DNase before proceeding with the column selection?
- Place on the 37 °C heat block for 30 min, and meanwhile prepare the columns.
- Immediately begin to prepare the spin columns - with all the incubations and spins, it will take longer than you think!
- During spins and incubations, you might also be able to begin Part 2 (steps 1+2) below.
- When the 30 min incubation is over, place your digested IVTs on ice. Cool for at least 2 min before putting the RNA onto the spin columns.
- Take one Micro Bio-Spin Column and one 2 mL microfuge tube per reaction, and for each:
- Rapidly flip the column back and forth a few times to resuspend the gel, then flick or tap it to rid air bubbles.
- Snap off the plastic tab at the bottom of the column (a little buffer may flow out), and place the column in the 2 mL microfuge tube. Remove the cap and let the excess buffer drain off.
- This will take just a few minutes. The resin should settle to about 1-2 mm above the narrowed neck of the column.
- If the buffer won't start flowing, talk to the teaching faculty. Sometimes pushing the cap on and then off will help it begin. Don't repeat this action over and over, though, or you may introduce air bubbles into the column.
- When the buffer stops flowing, discard the eluant into a waste tube and centrifuge the tube for 2 min at 1000 g. (No need to replace the cap.)
- Is g the same as rcf or rpm? Ask if you’re not sure!
- Add 500 μL of selection buffer (SB) to the top of the column. Let it drain by gravity for a couple of minutes, then spin for 1 min at 1000 g.
- You don't need to drain most of the fluid this time, just a few drops. However, if you skip immediately to centrifuging, the resin will retain a lot of extra buffer and dilute your RNA.
- Repeat step 7 three more times. Meanwhile, label two 1.5 mL eppendorf tubes for collection (on the side and with a sticky label on the cap indicating the RNA sequence identity), and cut off their caps.
- Transfer each column to a 1.5 mL tube – both column and tube should be labeled, to be on the safe side – then add 75 μL of your IVT reaction gently to the center of the top surface of the gel.
- Centrifuge for 4 min, at 1000 g. Carefully replace the cap on each sample, and store on ice for now.
- Hand in your used polyacrylamide columns up front so we can dispose of them in the solid waste container in the hood.
Part 2: Quantify RNA recovery
- For each RNA sample, you will add 5 μL of RNA to 695 μL of water. This ought to give you a measurement that's in a reliable range. You can prepare labeled eppendorf tubes containing the appropriate amount of water in advance.
- To get the most accurate measurement, you should also prepare a blanking solution that contains everything except the RNA, i.e., 695 μL of water and 5 μL of selection buffer.
- After eluting your RNA samples, pipet each gently and briefly to mix (avoid making bubbles!), then take 5 μL of the RNA and add to a water-filled eppendorf.
- Add 690 μL of your three solutions to separate cuvettes and head to the spec.
- Notice that today we are using cuvettes made of a special plastic that is transparent to UV light.
- Begin with the cuvette containing blanking solution, and hit Blank on the spectrophotometer.
- Proceed to take an absorbance scan of each RNA sample. Record the 260 nm and 280 nm absorbance values in your notebook. You can simply touch your finger to the onscreen spectrum for coarse wavelength selection, then touch the onscreen arrows for fine selection.
- What assumption are we making about the cuvettes when we put the blanking solution in a separate cuvette from the samples?
- Input your 260 and 280 nm data into the Excel sheet that you prepared for homework.
- Recall that an absorbance value of 1 indicates a concentration of 40 μg/mL of the measured RNA (i.e., the diluted solution and not the original stock).
- Which of the two wavelengths indicates RNA quantity? Re-read the introduction if you're not sure.
- Recall that an absorbance value of 1 indicates a concentration of 40 μg/mL of the measured RNA (i.e., the diluted solution and not the original stock).
Part 3: Prepare RNA for selection
- Dilute 75 μL of each RNA sample in selection buffer (not water!) to a concentration of 8 μM.
- Note that 85-90 μL of RNA is usually recovered after column elution. If you have <75 μL, let the teaching faculty know.
- The respective molecular weights of the RNAs are 31,344 g/mol and 33, 824 g/mol for 6-5 and 8-12.
- Most likely your final volume will be 0.5-1.5 mL; check with the teaching faculty if not.
- Before continuing on, make sure you have enough of each putative aptamer to follow through on your experimental plan. If not, talk to the teaching faculty.
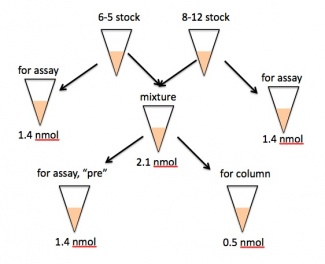
- Prepare 2.1 nmol of your 6-5/8-12 aptamer mixture.
- For example, a 20% 8-12 mixture would contain 0.42 nmol of 8-12 and 1.68 nmol of 6-5 RNA.
- Set aside 1.4 nmol of the mixture in a well-labeled eppendorf tube - this will serve as your pre-selection sample in the binding assay on Day 7 and should be kept on ice for now.
- Another 0.5 nmol of the mixture will be used for the two column selections (0.25 nmol each). Add 178 μL of selection buffer to 0.5 nmol of RNA. This mixture should be denatured at 70 °C for 5 min, then cooled at room temperature for at least 10 min.
- After the cooling period, add 160 μL of 125 μg/mL tRNA to your sample and pipet gently to mix.
- What is the final concentration of tRNA in your sample?
- Retrieve two 1 mL aliquots of hemin bead slurry from the teaching faculty. Spin each one for 1 min at 1000 g, and remove the ~900 μL of supernatant by pipetting.
- The beads have been washed and incubated with selection buffer for you.
- Finally, add half of your RNA sample to each tube of beads, cap tightly, cover with foil, and put it on the nutator for 1 hour.
- You should have ~400 μL of RNA total, but due to volume losses you might add slightly less than 200 μL to each tube, say 190-195 μL.
- As your benchmarks for the binding assay, you should also set aside exactly 1.4 nmol each of 6-5 and 8-12 alone. Give these two eppendorfs, along with the "pre" sample, to the teaching faculty, making sure they are well-labeled and tightly capped. These samples will be stored at -80 °C until needed. Please also hand over any extra 6-5 and 8-12, in case it is needed by one of your peers.
- If you are concerned about evaporation, and have sample to spare, you may set aside more than 1.4 nmol if you wish. Just remember to take only 1.4 nmol for testing on the binding assay day.
During the one hour incubation, you may write in your notebook, leave lab for a snack break, prepare for Part 4 below, or otherwise spend the time as you see fit.
Part 4: Column selection
- You and your partner can share one ring-stand. Affix two columns to the ring-stand such that you each have access to one of them. (See picture.)
- Snap the bottom off of each column. Save the top and bottom caps for later use.
- Wet each column with 200 μL of SB, and let it drain.
- Add one tube of resin/aptamer mixture to each column, and let drain.
- You may need to adjust your pipetting pace to get a uniform suspension of beads into the tip.
- Now you must rinse non-binding materials from the column. Start by adding 200 μL of SB to the top of the column, and allow it to drain.
- Repeat X times, according to the wash protocol you signed up for. Make sure each of you knows which partner is doing which wash number! Each wash should only take a few seconds.
- Finally, plug up the bottom of the column using the yellow cap, and add 200 μL of 2.5 mM hemin, which will be used to elute the aptamers. Incubate for 10 min. Loosely place the top cap on the column during this incubation.
- Uncap the column, and allow the eluant to drain by gravity into a well-labeled eppendorf.
Part 5: Begin RNA precipitation
- Measure the approximate volume (to within a few μL) of solution recovered.
- Add 1/10 volume of ammonium acetate, 1/200 V glycogen, and 2.5 V 100% ethanol, in that order.
- For example, for a 200 μL solution, you would add 20 μL of the ammonium acetate.
- Give the teaching faculty your samples to be put away at -20 °C until next time.
For next time
Parts 1, 2, and 3 of this assignment are also due on Stellar.
- Write up the next part of your Methods section (Days 3+4 of the module), applying the feedback you received on your previous draft.
- If you choose to re-write Days 1+2, you will get feedback on the success of that revision, but your grade will be based on Days 3+4.
- Write the opening context-setting paragraph of your Results section, in which you introduce the system you are working with and your immediate investigative goal.
- Draft an outline of the rest of the results to indicate the approximate high-level content and its order. You don't need to state the actual results, just each type of result that will be discussed in about a sentence each. Essentially, what order will your figures/tables/data be in and what will they consist of? Also include sub-section titles.
- Prepare the experimental schematic that will be included either in your Introduction or at the beginning of the Results.
- Recall that our collaboration and integrity guidelines state that your text and figures must be your own unless otherwise specified. Thus, you should all make your own schematic rather than modifying one from class slides. Keep in mind that you are being evaluated not on advanced PowerPoint skills, but instead on showing that you understand the purpose and major steps of the experiment and can convey them.
Reagent list
- DNase stock concentration is 2000 U/mL (from New England Biolabs)
- Micro Bio-Spin Columns
- Resin: Bio-Gel P30 in Tris buffer
- 40,000 MW cut-off
- RNase free
- From Bio-Rad
- Selection Buffer (SB)
- 100 mM Tris-acetate
- 500 mM Na-acetate
- 25 mM K-acetate
- 10 mM Mg-acetate
- 5% DMSO
- 0.05 % Triton-X
- Poly-Prep Chromatography Columns from Bio-Rad
- Yeast tRNA
- From Invitrogen
- 125 μg/mL in SB
- Hemin-agarose beads
- Distributed as 1 mL of 10X slurry (1 part resin: 9 parts solution)
- 4 μmoles hemin per mL resin
- From Sigma-Aldrich
- Pure hemin
- Solid chemical stock from Sigma-Aldrich
- Concentrated stock solution (~ 10-15 mM) prepared in DMSO
- Diluted to 2.5 mM in SB
- RNA Precipitation (stock solutions)
- 5 M ammonium acetate
- 20 mg/mL glycogen