20.109(F16):Prepare comet chips (Day1)
Contents
Introduction
Most of the laboratory exercises you will complete for this module are based on the CometChip assay developed in the Engelward Laboratory. The CometChip assay is used to assess various types of DNA lesions, including base excisions, abasic sites, strand breaks, and crosslinks.
To measure DNA damage, the CometChip assay relies on gel electrophoresis. Electrophoresis is a technique used to separate molecules by size using an applied electrical field and a sieving matrix. DNA, RNA and proteins are often studied with this technique; agarose and acrylamide gels are the two most common sieves. The molecules to be separated enter the matrix through a well at one end and are pulled through the matrix when a current is applied. Because DNA and RNA are negatively charged molecules due to their phosphate backbone, they naturally travel toward the positive charge at the far end of the gel. Larger molecules are entwined in the matrix and are stalled; smaller molecules wind through the matrix more easily and travel further from the well. Over time fragments of similar length accumulate into 'bands' in the gel. The CometChip assay is based upon the principle that damaged DNA travels more readily compared to undamaged DNA.
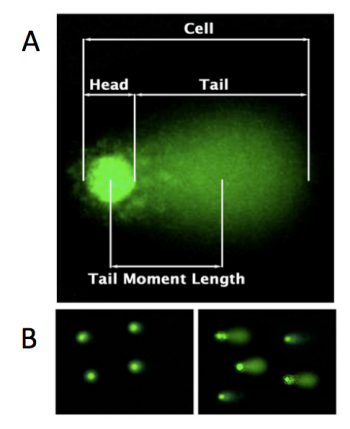
In the CometChip assay, cells are loaded into microwells that are 'stamped' onto an agarose gel sieving matrix. Specific treatments can then be applied to the cells within these microwells that induce DNA damage. The cells are then lysed to release the DNA into the microwell. Following cell lysis, the CometChip is incubated in an alkaline buffer that unwinds the DNA. This step allows for all types of DNA damage to be detected. Lastly, gel electrophoresis is used to separate the DNA fragments. DNA fragments migrate away from the microwell and generate a comet tail as shown in the image to the right (panel A). The distance that the DNA migrates (i.e. the length of the comet tail) is proportional to the amount of damage. For example, with cells not treated with a DNA damaging agent, there are no evident comet tails to the right of the microwell or 'head' (panel B, image on the left); however, in cells treated with a DNA damaging agent there are comet tails apparent (panel B, image on the right). Comet tail lengths can be compared across experimental treatments to determine the deleterious effects of chemicals and toxins on DNA stability. In addition, the CometChip assay can be used to study the rate and efficiency of repair in response to specific treatments.For this module you will complete several CometChip experiments in an effort to optimize the assay. In these experiments, the comet tails will be visualized using fluorescence microscopy and analyzed with code written by members of the Engelward Laboratory. Today you will complete the first steps of the CometChip assay by generating an agarose CometChip and preparing the cells that you will load into the microwells of your CometChip during the next laboratory meeting.
Protocols
Part 1: Laboratory orientation quiz
Complete the orientation quiz with your partner. Though you are working with your partner, each student should record all answers on the provided quiz. If you disagree with your partner on an answer, you should write what you think is the correct answer on your quiz.
Good luck!
Part 2: Prepare CometChip
- Obtain a sheet of gelbond film from the laboratory bench at the front of the room. The paper is protecting the hydrophilic side of the gelbond film.
- Be sure to keep the paper associated with the gelbond film so you know which side is which.
- Use the ruler in your team drawer to draw a 5 x 9 cm rectangle on the (hydrophobic side of the) gelbond film.
- Note: you are writing on what will be the bottom of the CometChip and may want to write backwards so the labels are clear when you look at the top of your CometChip.
- Prepare 20 mL of 1% normal melting point (NMP) agarose. Be careful as the agarose solution will be very hot!
- Calculate the amount of NMP agarose powder needed for a 1% w/v solution. Check your math with the teaching faculty before you continue.
- Obtain a small milk bottle from the front bench.
- Weigh out the appropriate amount of NMP agarose and add it to the milk bottle.
- Use a cylinder to measure 20 mL of 1x PBS and add it to the milk bottle with the NMP agarose powder.
- Swirl to mix.
- To melt the NMP agarose, microwave the solution for 20 seconds, swirl, then microwave for 3-second intervals until all crystals are in solutions. After each interval, remove the milk bottle and gently swirl while checking for unmelted agarose crystals. It is important that the solution does NOT boil as you will lose water to evaporation and the density of the agarose will be altered. If your solution starts to boil, immediately remove it from the microwave and gently swirl.
- When no more crystals are visible in the solution take the milk bottle to your bench.
- Obtain a small rectangle dish and the CometChip 'stamp' from the front bench.
- Add 2.5 mL of the agarose solution to the small dish, then quickly place the gelbond film in the dish with the marked hydrophobic side down.
- Use a P1000 blue pipet tip to gently tap the gelbond film in order to remove any air bubble as well as ensure that it lays very flat in the dish.
- Add 13 mL of the agarose solution on top of the gelbond film.
- Slowly place the CometChip stamp on top of the agarose.
- Lower the bottom left of the stamp first, then slowly allow the stamp to 'roll' into the agarose. Be sure to leave the top right corner of the small dish accessible.
- Be careful not to introduce bubbles into the agarose and work quickly as the agarose will solidify as it cools.
- Allow the agarose to solidy, undisturbed, on your bench for 30 min.
- Add ~5 mL of 1x PBS to the small dish that contains your agarose CometChip.
- Pipet in the 1x PBS using the accessible corner.
- Slowly pull from one corner of the stamp to lift it away from your CometChip in the dish.
- If the CometChip sticks to the stamp, carefully peel it off using tweezers.
- Discard the PBS in the sink.
- Remove excess agarose from the perimeter of your CometChip using a razor blade.
- Clean the agarose from the bottom of your CometChip (gelbond side) using a Kimwipe.
- Place your CometChip in the small dish containing 1x PBS for storage at 4 °C until next time.
- Be sure the chip is completely submerged.
Part 3: Split mammalian cells
In the past century, we have learned a tremendous amount by studying the behavior of mammalian cells maintained in the laboratory. Tissue culture was originally developed about 100 years ago as a method for learning about mammalian biology. The term tissue culture was coined because people were doing exactly that, extracting tissue and letting it live in a dish for a short time. Today, most tissue culture experiments are done using isolated cells rather than whole tissues. Much of what we know about cancer, heritable diseases, and the effects of the environment on human health has been derived from studies of cultured cells.
Cells that are isolated from tissue are called primary cells, because they come directly from an animal. It is very difficult to culture primary cells, largely because primary cells that are placed in culture divide only a limited number of times. This limitation on the lifespan of cultured primary cells, called the Hayflick limit, is a problem because it requires a researcher to constantly remove tissues from animals in order to complete a study. Cell isolation processes can be quite labor-intensive, and also can complicate data analysis due to inherent animal-to-animal variation. To get around the first of these problems, researchers use cells that are immortal, which means they can divide indefinitely, though some inherent cell-to-cell variation still exists in such cells.
One familiar type of immortalized cell is the cancer cell. Tumor cells continuously divide, allowing cancer to invade tissues and proliferate. Cancer cells behave the same way in culture, and under the right conditions, cells can be taken from a tumor and divide indefinitely in culture. Another type of immortalized cell is the embryonic stem cell. Embryonic stem cells are derived from an early stage embryo, and these cells are completely undifferentiated and pluripotent, which means that under the right conditions, they can become any mammalian cell type.
The art of tissue culture lies in the ability to create conditions that are similar to what a cell would experience in an animal, namely 37 °C and neutral pH. Blood nourishes the cells in an animal, and blood components are used to feed cells in culture. Serum, the cell-free (and clotting-factor free) component of blood, contains many of the factors necessary to support the growth of cells outside the animal. Consequently, serum is frequently added to tissue culture medium, although serum-free media exist and support some types of cultured cells. Furthermore, cultured mammalian cells must grow in a germ-free environment and researchers using tissue culture must be skilled in sterile technique.
One major objective for this experimental module is for you to learn how to perform tissue culture using several cell lines. In this specific experiment, you will use TK6 cells. The biological source of these cells is a lymphoblast from a human host.
- Clean the tissue culture hood and prepare it with the supplies you will need according to the demonstration provided by the teaching faculty during the Orientation exercise.
- Read through the protocol to ensure you have all of the equipment needed.
- Obtain a ~48 h culture of TK6 cells in a T75 flask from the 37 °C incubator.
- Determine the density of the TK6 culture (cells per mL) using a hemocytometer as done previously during the Orientation exercise. As a brief reminder:
- Transfer 90 μL of the TK6 cell culture to a 1.5 mL eppendorf tube.
- Carry the tube to the center microscope bench and add 10 μL of trypan blue cell stain.
- Carefully pipet 10 μL of the stained cells between the hemocytometer and glass cover slip.
- Count the cells that fall within the corner squares of the 4x4 pattern, average (i.e. divide by 4), and then multiply by 10,000 to determine the number of cells/mL.
- Calculate the volume of cell culture needed to seed with 2,000,000 cells.
- Obtain a sterile T75 flask and an aliquot of pre-warmed fresh tissue culture media.
- Clearly label your T75 flask with the date, cell line, and your team information.
- Transfer the volume of cell culture you calculated in Step #4 from the T75 flask containing the ~48 hr TK6 culture to the sterile T75 flask.
- Add enough fresh tissue culture media to your new T75 flask such that the total volume is 12 mL.
- Move your new TK6 cell culture to the 37 °C incubator.
- Be sure to place your flask on the appropriate shelf.
- Clean out the tissue culture hood according to the demonstration provided by the teaching faculty during the Orientation exercise.
Reagents list
CometChip:
- agar, normal melting point (Invitrogen)
- phosphate buffered saline
- GelBond film (Lonza)
- 1 well dish (VWR)
Cell culture:
- RPMI 1640 (Invitrogen)
- with 10% fetal bovine serum (Atlanta Biologicals)
- 100X antibiotic solution (Invitrogen)
- 10,000 U/mL Penicillin
- 10,000 U/mL Streptomycin
- Trypan Blue
- Incubator maintains 37°C, 5% CO2 and 95% relative humidity
Next day: Develop experiment to test loading variables and quantify growth rate
Previous day: Orientation