Difference between revisions of "Spring 2012:Leanna Morinishi Lab 1"
(→Methodology) |
(→Methodology) |
||
| Line 38: | Line 38: | ||
<math> F = \frac{k_{B}T}{l_{p}}[(\frac{1}{4}(1 - \frac{x}{l_{c}})^{-2} - \frac{1}{4} +\frac{x}{l_{c}}]</math><br/> | <math> F = \frac{k_{B}T}{l_{p}}[(\frac{1}{4}(1 - \frac{x}{l_{c}})^{-2} - \frac{1}{4} +\frac{x}{l_{c}}]</math><br/> | ||
| − | where k_{B} is the Boltzmann constant, l_{c} is the contour length and l_{p} is the persistence length. | + | where <math>k_{B}</math> is the Boltzmann constant, <math> l_{c}</math> is the contour length and <math> l_{p}</math>is the persistence length. |
== Results == | == Results == | ||
Revision as of 15:54, 28 February 2012
Lab 1: Optical Trapping
Goals
We set out to add functionality to the trap control software, specifically we wanted to automate centering of the DNA-tethered bead using a centering algorithm which we would write in MATLAB. To center the DNA-tethered bead we would need to learn how to communicate with different electronics using DAQ and the daqtoolbox in MATLAB, how to test a code which is supposed to provide specific functionality, and a bit about the UI in MATLAB. After writing the code, our goals expanded to include writing code to simulate a typical optical trap experiment. Beyond this, we tried to:
- Get nicer calibration data
- Look at the properties of the tether, to calculate the persistence and contour lengths
Skills Learned
- How to talk to the DAQ and ActiveX controls
- More about QPD and piezo-electric
- How to put a button on a UI in Matlab!!
Methodology
Materials and Instruments
- Matlab: programming language
- DAQ
- Piezoelectric Stages
- Bead size: 0.97 um beads used
- Laser wavelength: λ = 975 nm
Centering Function
- First, I learned how to activate channels, and talk to the DAQ
- I created a sine wave and had it repeat infinitely, then acquired that same data from the DAQ
- In the actual code, we decided to integrate into OTKB.m, so the following is just the functions within that code
- We begin by establishing a waveform in the x-direction, calculating the center position
- Repeat in y-direction, then x-direction 4 times
- Take the mean calculated center positions and use that as the final centering position
- Invoke piezos to reset the center voltage at our calculated position
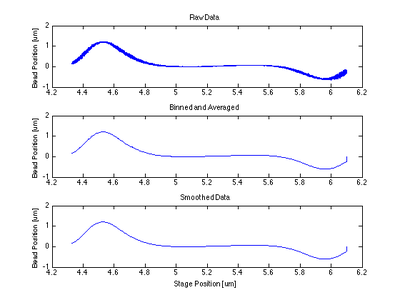
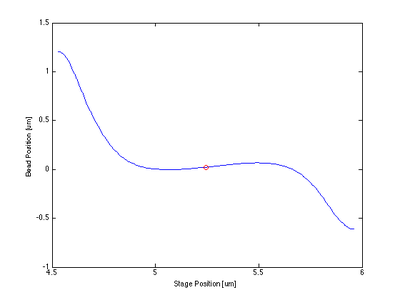
- The centering function itself required only that I bin and average my data, smooth it, then find the indices of the max and min and take information at the point between them.
Estimating contour and persistence lengths
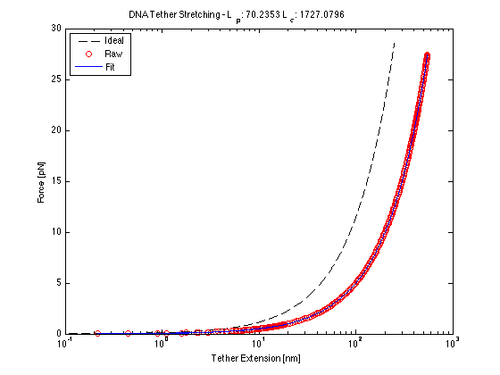
The worm-like chain model is one method to approximate the behavior of DNA when stretched. Here we are interested in the Extension curve of a DNA tether, so that we may use its (hypothetically) linear region to calculate the place of tether attachment. We fitted our data (guided by calibration calculations) to the following model equation using nlinfit to solve for the persistence and contour lengths.
$ F = \frac{k_{B}T}{l_{p}}[(\frac{1}{4}(1 - \frac{x}{l_{c}})^{-2} - \frac{1}{4} +\frac{x}{l_{c}}] $
where $ k_{B} $ is the Boltzmann constant, $ l_{c} $ is the contour length and $ l_{p} $is the persistence length.
Results
Force/Stretching Estimation using nlinfit
(Calculated contour and persistence lengths are in nm at 20mW)
Proof of Concept for Center Calculation
The following data is one example, taken from tethered bead data at 20 mW.
Code
Centering Function
function pushbuttonDNATetherCenter_Callback(hObject, eventdata, handles)
dnaTetherCentering(handles, uiSettings)
function DNATetherCentering(handles, uiSettings)
accuracy = 1;
accuracyY = 1;
setParams = false;
xcenter = false;
ycenter = false;
while accuracy < 4;
if ~setParams
xaxisPiezoDriver = handles.PiezoDriverDescriptorList{1};
yaxisPiezoDriver = handles.PiezoDriverDescriptorList{2};
handles.SamplesToSave = uiSettings.numberofSeconds*uiSettings.sampleRate;
Amplitude = uiSettings.stageOscillationAmplitude;
waveformFreq = round(uiSettings.sampleRate / uiSettings.stageOscillationFrequency);
time = linspace(0:length(numberOfSamples)-1,numberOfSamples);
centeringcycle = Amplitude*sin(2*pi*waveformFreq * time)';
waveform = [centeringcycle zeros(length(centeringcycle),1)];
numberOfSamples = sampleRate * duration; %set sampleRate and duration;
set(handles.DaqInput.ObjectHandle,'SampleRate',400);
set(handles.DaqInput.ObjectHandle,'Trigger','Manual');
set(handles.DaqInput.ObjectHandle,'SamplesPerTrigger',numberOfSamples);
set(handles.DaqOutput.ObjectHandle,'Trigger','Manual');
set(handles.DaqOutput.ObjectHandle,'RepeatOutput',3);
%
setParams = true;
end
start(handles.DaqInput.ObjectHandle);
data = getdata(handles.DaqInputHandle);
Trigger(handles.DaqInput.Output.ObjectHandle);
putdata(handles.DaqOutput.ObjectHandle,waveform);
start(handles.DaqOutput.ObjectHandle);
Trigger(handles.DaqOutput.ObjectHandle);
waveform = findCenter(data, xcenter, ycenter, waveform);
stop(handles.DaqInput.ObjectHandle);
stop(handles.DaqOutput.ObjectHandle);
end
function waveform = findCenter(data, xcenter, ycenter, waveform)
[quantizedXAxisx BinnedDatax StandardDeviationx Countx] = BinData( ...
data, 'XColumn', 3, 'YColumn', 1);
[quantizedXAxisy BinnedDatay StandardDeviationy County] = BinData( ...
data, 'XColumn', 4, 'YColumn', 2);
data = [BinnedDatax' BinnedDatay' quantizedXAxisx' quantizedXAxisy']; % qpdx qpdy piezx piezy
centeredPosition = (positionOfMaxVoltage + positionOfMinVoltage)/2;
fprintf('The position of the stage is %d',centeredPosition);
if ~xcenter && ~ycenter
[MaxValue MaxVoltageIndex] = max(data(:,1));
positionOfMaxVoltage = centeringcycle(MaxVoltageIndex(1));
[MinValue MinVoltageIndex] = min(data(:,1));
positionOfMinVoltage = centeringcycle(MinVoltageIndex(1));
centeredPosition = (positionOfMaxVoltage + positionOfMinVoltage)/2;
invoke(handles.PiezoDriverDescriptorList{1}.DriverActiveXControl,...
'SetPosOutput', 0, centeredPosition);
xcenter = true;
waveform = [zeros(length(centeringcycle),1) centeringcycle ];
elseif xcenter && ~ycenter
[MaxValue MaxVoltageIndex] = max(data(:,2));
positionOfMaxVoltage = centeringcycle(MaxVoltageIndex(1));
[MinValue MinVoltageIndex] = min(data(:,2));
positionOfMinVoltage = centeringcycle(MinVoltageIndex(1));
centeredPosition = (positionOfMaxVoltage + positionOfMinVoltage)/2;
invoke(handles.PiezoDriverDescriptorList{2}.DriverActiveXControl,...
'SetPosOutput', 0, centeredPosition);
fprintf('The position of the stage is %d',centeredPosition);
waveform = [ centeringcycle zeros(length(centeringcycle),1)];
if accuracyY == 4;
centeredPosition = mean(CheckaccuracyYposition);
invoke(handles.PiezoDriverDescriptorList{2}.DriverActiveXControl,...
'SetPosOutput', 0, centeredPosition);
end
CheckaccuracyYposition(accuracyY) = centeredPosition;
accuracyY = accuracyY + 1;
ycenter = true;
elseif xcenter && ycenter
[MaxValue MaxVoltageIndex] = max(data(:,1));
positionOfMaxVoltage = centeringcycle(MaxVoltageIndex(1));
[MinValue MinVoltageIndex] = min(data(:,1));
positionOfMinVoltage = centeringcycle(MinVoltageIndex(1));
centeredPosition = (positionOfMaxVoltage + positionOfMinVoltage)/2;
CheckaccuracyXposition(accuracy) = centeredPosition;
waveform = [ centeringcycle zeros(length(centeringcycle),1)];
% center to mean value of all checkaccuracyXposition
if accuracy == 4;
centeredPosition = mean(CheckaccuracyXposition);
invoke(handles.PiezoDriverDescriptorList{1}.DriverActiveXControl,...
'SetPosOutput', 0, centeredPosition);
end
accuracy = accuracy +1;
ycenter = false;
end
Persistence and Contour Length Estimation
Y = 1; % Young's modulus
I = 1; % moment of inertia
T = 293; % Temperature K
k_B = 1.38e-23; % Boltzmann's constant (m^2 kg)/(s^2 K)
x = 10; % end to end extension of DNA tether
l_p = 45; % persistence length nm (Y * I)/(k_B * T);
l_c = 1180; % contour length nm
datax = load('pretty nice DNA tether 20mw 1.txt');
datax(:,3) = datax(:,3)*2.22; % 2.22 um/V
datax(:,1) = datax(:,1)/(.5); % .5 V/um
location = [1 1];
edgesx = linspace(min(datax(:,3)), max(datax(:,3)), 1e3);
[l, whichbinx] = histc(datax(:,3), edgesx);
binmeansx = zeros(1, length(edgesx-1));
for i = 1:length(edgesx)-1
flagmembers = (whichbinx == i);
members = datax(flagmembers,1);
binmeansx(i) = mean(members);
end
smoothedx = smooth(binmeansx,200);
[xmin xminindex] = min(smoothedx);
[xmax xmaxindex] = max(smoothedx);
middlex = ceil(mean([xminindex xmaxindex]));
x_stage = edgesx(xmaxindex:xminindex); %, smoothedx(xmaxindex:xminindex)
x_bead = 1e3*abs(smoothedx(middlex) - smoothedx(xmaxindex:xminindex))';
y = x_bead*5e-5; % 5e-5 N/um
myFunction = @ (x, xdata) forceApplied(Y, I, T, x_bead, x(1), x(2));
beta = nlinfit(x_bead, y, myFunction, [45, 1180]);
figure()
semilogx(x_bead, forceApplied(Y, I, T, x_bead, beta(1), beta(2)))
ylabel('Force [pN]')
xlabel('Tether Extension [nm]')
title(['DNA Tether Stretching - L_p: ', num2str(beta(1)), ' L_c: ', num2str(beta(2))])</nowiki