Difference between revisions of "Spring 2012:Leanna Morinishi Lab 1"
From Course Wiki
(→Proof of Concept for Center Calculation) |
(→How I did it) |
||
| Line 13: | Line 13: | ||
* How to put a button on a UI in Matlab!! | * How to put a button on a UI in Matlab!! | ||
| − | == How | + | == How We did it == |
=== Centering Function === | === Centering Function === | ||
* First, I learned how to activate channels, and talk to the DAQ | * First, I learned how to activate channels, and talk to the DAQ | ||
| Line 24: | Line 24: | ||
* The centering function itself required only that I bin and average my data, smooth it, then find the indices of the max and min and take the point in the middle of them. | * The centering function itself required only that I bin and average my data, smooth it, then find the indices of the max and min and take the point in the middle of them. | ||
| + | |||
| + | === Estimating contour and persistence lengths === | ||
| + | |||
| + | <math> F = \frac{k_{B}}{l_{p}}[(\frac{1}{4}(1 - \frac{x}{l_{c}})^{-2} - \frac{1}{4} +\frac{x}{l_{c}}]</math><br/> | ||
| + | |||
| + | where k_{B} is the Boltzmann constant, l_{c} is the contour length and l_{p} is the persistence length. | ||
== Results == | == Results == | ||
Revision as of 08:46, 28 February 2012
Contents
Lab 1: Optical Trapping
What I wanted to accomplish
- Complete a button that takes in data from a tethered microbead and adjusts the stage position to center it on the tether
- Try to get nicer calibration data
- Function that finds the center
- Feed in recorded data in a simulation
- Look at the properties of the tether, to calculate the persistence and contour lengths
What I have learned
- How to talk to the DAQ and ActiveX controls
- More about QPD and piezo-electric
- How to put a button on a UI in Matlab!!
How We did it
Centering Function
- First, I learned how to activate channels, and talk to the DAQ
- I created a sine wave and had it repeat infinitely, then acquired that same data from the DAQ
- In the actual code, we decided to integrate into OTKB.m, so the following is just the functions within that code
- We begin by establishing a waveform in the x-direction, calculating the center position
- Repeat in y-direction, then x-direction 4 times
- Take the mean calculated center positions and use that as the final centering position
- Invoke piezos to reset the center voltage at our calculated position
- The centering function itself required only that I bin and average my data, smooth it, then find the indices of the max and min and take the point in the middle of them.
Estimating contour and persistence lengths
$ F = \frac{k_{B}}{l_{p}}[(\frac{1}{4}(1 - \frac{x}{l_{c}})^{-2} - \frac{1}{4} +\frac{x}{l_{c}}] $
where k_{B} is the Boltzmann constant, l_{c} is the contour length and l_{p} is the persistence length.
Results
Proof of Concept for Center Calculation
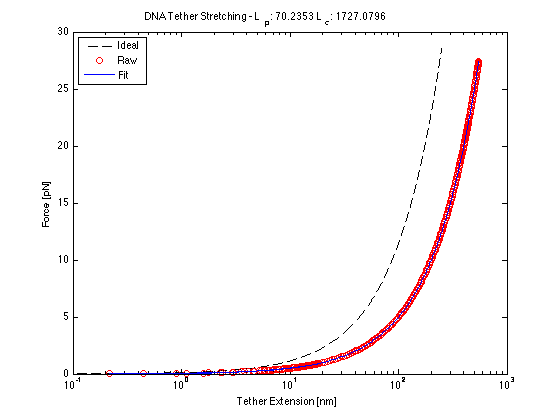
The following data is one example, taken from tethered bead data at 20 mW.
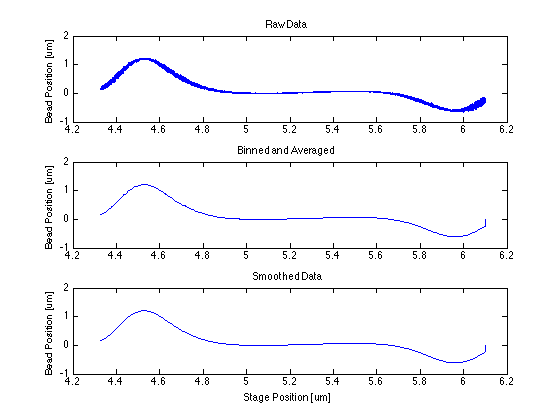
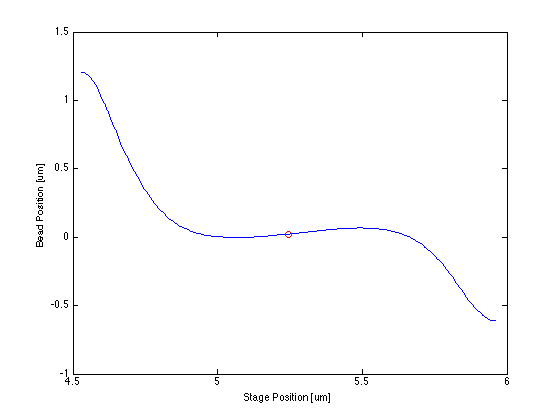
Force/Stretching Estimation using nlinfit
(Calculated contour and persistence lengths are in nm at 20mW)
Code
Centering Function
function pushbuttonDNATetherCenter_Callback(hObject, eventdata, handles)
dnaTetherCentering(handles, uiSettings)
function DNATetherCentering(handles, uiSettings)
accuracy = 1;
accuracyY = 1;
setParams = false;
xcenter = false;
ycenter = false;
while accuracy < 4;
if ~setParams
xaxisPiezoDriver = handles.PiezoDriverDescriptorList{1};
yaxisPiezoDriver = handles.PiezoDriverDescriptorList{2};
handles.SamplesToSave = uiSettings.numberofSeconds*uiSettings.sampleRate;
Amplitude = uiSettings.stageOscillationAmplitude;
waveformFreq = round(uiSettings.sampleRate / uiSettings.stageOscillationFrequency);
time = linspace(0:length(numberOfSamples)-1,numberOfSamples);
centeringcycle = Amplitude*sin(2*pi*waveformFreq * time)';
waveform = [centeringcycle zeros(length(centeringcycle),1)];
numberOfSamples = sampleRate * duration; %set sampleRate and duration;
set(handles.DaqInput.ObjectHandle,'SampleRate',400);
set(handles.DaqInput.ObjectHandle,'Trigger','Manual');
set(handles.DaqInput.ObjectHandle,'SamplesPerTrigger',numberOfSamples);
set(handles.DaqOutput.ObjectHandle,'Trigger','Manual');
set(handles.DaqOutput.ObjectHandle,'RepeatOutput',3);
%
setParams = true;
end
start(handles.DaqInput.ObjectHandle);
data = getdata(handles.DaqInputHandle);
Trigger(handles.DaqInput.Output.ObjectHandle);
putdata(handles.DaqOutput.ObjectHandle,waveform);
start(handles.DaqOutput.ObjectHandle);
Trigger(handles.DaqOutput.ObjectHandle);
waveform = findCenter(data, xcenter, ycenter, waveform);
stop(handles.DaqInput.ObjectHandle);
stop(handles.DaqOutput.ObjectHandle);
end
function waveform = findCenter(data, xcenter, ycenter, waveform)
[quantizedXAxisx BinnedDatax StandardDeviationx Countx] = BinData( ...
data, 'XColumn', 3, 'YColumn', 1);
[quantizedXAxisy BinnedDatay StandardDeviationy County] = BinData( ...
data, 'XColumn', 4, 'YColumn', 2);
data = [BinnedDatax' BinnedDatay' quantizedXAxisx' quantizedXAxisy']; % qpdx qpdy piezx piezy
centeredPosition = (positionOfMaxVoltage + positionOfMinVoltage)/2;
fprintf('The position of the stage is %d',centeredPosition);
if ~xcenter && ~ycenter
[MaxValue MaxVoltageIndex] = max(data(:,1));
positionOfMaxVoltage = centeringcycle(MaxVoltageIndex(1));
[MinValue MinVoltageIndex] = min(data(:,1));
positionOfMinVoltage = centeringcycle(MinVoltageIndex(1));
centeredPosition = (positionOfMaxVoltage + positionOfMinVoltage)/2;
invoke(handles.PiezoDriverDescriptorList{1}.DriverActiveXControl,...
'SetPosOutput', 0, centeredPosition);
xcenter = true;
waveform = [zeros(length(centeringcycle),1) centeringcycle ];
elseif xcenter && ~ycenter
[MaxValue MaxVoltageIndex] = max(data(:,2));
positionOfMaxVoltage = centeringcycle(MaxVoltageIndex(1));
[MinValue MinVoltageIndex] = min(data(:,2));
positionOfMinVoltage = centeringcycle(MinVoltageIndex(1));
centeredPosition = (positionOfMaxVoltage + positionOfMinVoltage)/2;
invoke(handles.PiezoDriverDescriptorList{2}.DriverActiveXControl,...
'SetPosOutput', 0, centeredPosition);
fprintf('The position of the stage is %d',centeredPosition);
waveform = [ centeringcycle zeros(length(centeringcycle),1)];
if accuracyY == 4;
centeredPosition = mean(CheckaccuracyYposition);
invoke(handles.PiezoDriverDescriptorList{2}.DriverActiveXControl,...
'SetPosOutput', 0, centeredPosition);
end
CheckaccuracyYposition(accuracyY) = centeredPosition;
accuracyY = accuracyY + 1;
ycenter = true;
elseif xcenter && ycenter
[MaxValue MaxVoltageIndex] = max(data(:,1));
positionOfMaxVoltage = centeringcycle(MaxVoltageIndex(1));
[MinValue MinVoltageIndex] = min(data(:,1));
positionOfMinVoltage = centeringcycle(MinVoltageIndex(1));
centeredPosition = (positionOfMaxVoltage + positionOfMinVoltage)/2;
CheckaccuracyXposition(accuracy) = centeredPosition;
waveform = [ centeringcycle zeros(length(centeringcycle),1)];
% center to mean value of all checkaccuracyXposition
if accuracy == 4;
centeredPosition = mean(CheckaccuracyXposition);
invoke(handles.PiezoDriverDescriptorList{1}.DriverActiveXControl,...
'SetPosOutput', 0, centeredPosition);
end
accuracy = accuracy +1;
ycenter = false;
end
Persistence and Contour Length Estimation
Y = 1; % Young's modulus
I = 1; % moment of inertia
T = 293; % Temperature K
k_B = 1.38e-23; % Boltzmann's constant (m^2 kg)/(s^2 K)
x = 10; % end to end extension of DNA tether
l_p = 45; % persistence length nm (Y * I)/(k_B * T);
l_c = 1180; % contour length nm
datax = load('pretty nice DNA tether 20mw 1.txt');
datax(:,3) = datax(:,3)*2.22; % 2.22 um/V
datax(:,1) = datax(:,1)/(.5); % .5 V/um
location = [1 1];
edgesx = linspace(min(datax(:,3)), max(datax(:,3)), 1e3);
[l, whichbinx] = histc(datax(:,3), edgesx);
binmeansx = zeros(1, length(edgesx-1));
for i = 1:length(edgesx)-1
flagmembers = (whichbinx == i);
members = datax(flagmembers,1);
binmeansx(i) = mean(members);
end
smoothedx = smooth(binmeansx,200);
[xmin xminindex] = min(smoothedx);
[xmax xmaxindex] = max(smoothedx);
middlex = ceil(mean([xminindex xmaxindex]));
x_stage = edgesx(xmaxindex:xminindex); %, smoothedx(xmaxindex:xminindex)
x_bead = 1e3*abs(smoothedx(middlex) - smoothedx(xmaxindex:xminindex))';
y = x_bead*5e-5; % 5e-5 N/um
myFunction = @ (x, xdata) forceApplied(Y, I, T, x_bead, x(1), x(2));
beta = nlinfit(x_bead, y, myFunction, [45, 1180]);
figure()
semilogx(x_bead, forceApplied(Y, I, T, x_bead, beta(1), beta(2)))
ylabel('Force [pN]')
xlabel('Tether Extension [nm]')
title(['DNA Tether Stretching - L_p: ', num2str(beta(1)), ' L_c: ', num2str(beta(2))])</nowiki