Difference between revisions of "DNA Melting: Simulating DNA Melting - Intermediate Topics"
| Line 1: | Line 1: | ||
==Overview== | ==Overview== | ||
| − | In the DNA Melting Lab, the raw data you will record consists of two voltages as they vary over time. In order to estimate the melting temperature, these raw voltages will have to be converted to a melting curve, which gives the fraction of dsDNA as a function of temperature. | + | In the DNA Melting Lab, the raw data you will record consists of two voltages as they vary over time. In order to estimate the melting temperature, these raw voltages will have to be converted to a melting curve, which gives the fraction of dsDNA as a function of temperature. T<sub>m</sub> can be estimated directly from the melting curve or by finding the peak of the melting curve's derivative. |
Data analysis steps include: | Data analysis steps include: | ||
| Line 9: | Line 9: | ||
# Differentiate the resulting function. | # Differentiate the resulting function. | ||
| − | It will be essential to have your data analysis scripts working before you come to the lab. This tutorial will demonstrate how to simulate | + | It will be essential to have your data analysis scripts working before you come to the lab. This tutorial will demonstrate how to simulate the DNA melting experiment in Matlab. You can use the simulated dataset to help develop your data analysis scripts. |
===Before you begin=== | ===Before you begin=== | ||
| Line 16: | Line 16: | ||
*You may also find it helpful to review the [[DNA Melting Thermodynamics|DNA Melting Thermodynamics Lecture Notes]]. | *You may also find it helpful to review the [[DNA Melting Thermodynamics|DNA Melting Thermodynamics Lecture Notes]]. | ||
| − | ==DNA melting | + | ==Simulate the DNA melting experiment== |
| − | + | ===Model the cooling of the sample=== | |
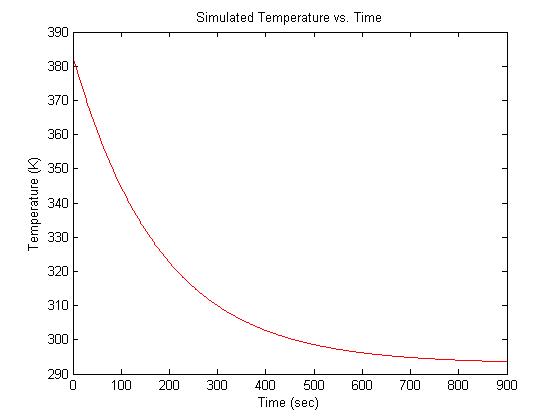
| + | At the beginning of an experimental run, the sample is heated in a bath to around 90°C. Over a period of about 15 minutes, the sample cools to room temperature of about 20°C. Cooling can be modeled by an exponential function with a time constant of 3 minutes. Thus, the equation for temperature (in °K) versus time is: | ||
| − | + | :<math> | |
| − | + | T(t) = 293 + 90 e ^ -\frac{t}{180} | |
| − | + | </math> | |
| − | + | ||
| − | + | In Matlab: | |
| + | <pre> | ||
| + | t = 1:899; % create a time vector (units: seconds) | ||
| + | T = 90 * exp(-1 * t / 180) + 293; % exponential cooling from 383K to 293K | ||
| + | plot(t,T); | ||
| + | </pre> | ||
| + | |||
| + | [[Image:DNA Sample Cooling.jpg]] | ||
| + | Remember that the semicolon at the end of the line prevents Matlab from displaying dozens of annoying lines on the console containing the value of t. | ||
==Write a function to compute f== | ==Write a function to compute f== | ||
Revision as of 04:22, 11 April 2008
Contents
Overview
In the DNA Melting Lab, the raw data you will record consists of two voltages as they vary over time. In order to estimate the melting temperature, these raw voltages will have to be converted to a melting curve, which gives the fraction of dsDNA as a function of temperature. Tm can be estimated directly from the melting curve or by finding the peak of the melting curve's derivative.
Data analysis steps include:
- Filter raw data to remove noise.
- Transform RTD voltage to temperature.
- Transform photodiode current to relative fluorescence.
- Ensure that the melting function is uniquely values by combining samples with identical temperature values.
- Differentiate the resulting function.
It will be essential to have your data analysis scripts working before you come to the lab. This tutorial will demonstrate how to simulate the DNA melting experiment in Matlab. You can use the simulated dataset to help develop your data analysis scripts.
Before you begin
- If you are need a review of Matlab fundamentals, see Matlab:Matlab Fundamentals
- Read the DNA Melting Lab Manual.
- You may also find it helpful to review the DNA Melting Thermodynamics Lecture Notes.
Simulate the DNA melting experiment
Model the cooling of the sample
At the beginning of an experimental run, the sample is heated in a bath to around 90°C. Over a period of about 15 minutes, the sample cools to room temperature of about 20°C. Cooling can be modeled by an exponential function with a time constant of 3 minutes. Thus, the equation for temperature (in °K) versus time is:
- $ T(t) = 293 + 90 e ^ -\frac{t}{180} $
In Matlab:
t = 1:899; % create a time vector (units: seconds) T = 90 * exp(-1 * t / 180) + 293; % exponential cooling from 383K to 293K plot(t,T);
Remember that the semicolon at the end of the line prevents Matlab from displaying dozens of annoying lines on the console containing the value of t.
Write a function to compute f
The first useful piece of code is a function that will compute $ \left . f \right . $ from the equation derived in class. This function must be in its own file called DnaFraction.m.
<include src="http://web.mit.edu/~20.309/www/Matlab/DnaFraction.m" />
%Returns the fraction of dsDNA given total DNA concentration, temperature, Delta S, and Delta H %Usage: f = DnaFraction(Ct, T, DeltaS, DeltaH) function f = DnaFraction(Ct, T, DeltaS, DeltaH) %Constants R=8.3; %first compute Ct * Keq CtKeq = Ct * exp(DeltaS / R - DeltaH / (R * T)); %now compute f f = (1 + CtKeq + sqrt(1 + 2 * CtKeq))/CtKeq;
Test the function
First, create a temperature vector. Then call DnaFraction with some reasonable parameters and plot the result. Units are calorie, mole.
t = [20:90] + 273; f = DnaFraction(.1E-6, t, 304E3, 786); plot(t,f)