Difference between revisions of "Lab Manual: Optical Microscopy"
(→=Bright field transmitted illumination imaging) |
|||
| Line 36: | Line 36: | ||
[[Image:20.309Hw3Imagelight.JPG|center|thumb|400px|Figure 1: Rough schematic showing white light and fluorescence light paths for imaging the specimen plane. Not drawn to scale.]] | [[Image:20.309Hw3Imagelight.JPG|center|thumb|400px|Figure 1: Rough schematic showing white light and fluorescence light paths for imaging the specimen plane. Not drawn to scale.]] | ||
| − | + | ==Microscopy lab etiquette== | |
| − | + | ||
| − | + | ||
| − | + | ||
* Observe all laser safety guidelines. | * Observe all laser safety guidelines. | ||
* Keep all of the boxes for the optics you use with your instrument to simplify putting things away. | * Keep all of the boxes for the optics you use with your instrument to simplify putting things away. | ||
| Line 49: | Line 46: | ||
* Use tube rings (never an SM1T2) to mount an optic in a lens tube. | * Use tube rings (never an SM1T2) to mount an optic in a lens tube. | ||
| − | + | ==Microscope components== | |
| − | + | ||
| − | + | ||
| − | + | ===Rigid optical construction=== | |
| + | Your design can use a combination to cage and lens tube components from ThorLabs. (See the [http://www.thorlabs.com/navigation.cfm?Guide_ID=50 ThorLabs online catalog] for more details. Print catalogs are available in the lab.) Be sure you understand how to use cage cubes ([http://www.thorlabs.com/thorProduct.cfm?partNumber=C4W C4W]), cube optic mounts ([http://www.thorlabs.com/thorProduct.cfm?partNumber=B5C B5C]), and kinematic mounting plates ([http://www.thorlabs.com/thorProduct.cfm?partNumber=B4C B4C]). Please ask about any components you are not sure how to use. | ||
| − | + | An example microscope put together by the instructors will be available in the lab for you to look at. Feel free to make improvements on this design. Stability will be crucial for the particle tracking experiments. This will be achieved through good design and careful construction — not by mindlessly overtightening screws. | |
| + | ===Simple lenses=== | ||
Plano-convex spherical lenses are available with focal lengths of 25, 50, 75, 100, 125, and 200 mm. It is best to mount most optics in short (0.5" or 0.3") lens tubes. It is acceptable to mount a lens between the end of a tube and a tube ring or between two tube rings. In most cases, the convex side of the lens faces toward the collimated beam; the planar side goes toward the convergent rays. Plano-concave lenses with focal lengths of -35 and -50 are also available. | Plano-convex spherical lenses are available with focal lengths of 25, 50, 75, 100, 125, and 200 mm. It is best to mount most optics in short (0.5" or 0.3") lens tubes. It is acceptable to mount a lens between the end of a tube and a tube ring or between two tube rings. In most cases, the convex side of the lens faces toward the collimated beam; the planar side goes toward the convergent rays. Plano-concave lenses with focal lengths of -35 and -50 are also available. | ||
| Line 65: | Line 62: | ||
Handle lenses only by the edges. If a lens is dirty, first remove grit with a blast of clean air or CO2. Clean the lens by wiping with a folded piece of lens paper wetted with a drop of methanol. (Do not touch the part of the tissue you use for cleaning with your fingers.) In some cases, it may be helpful to hold the folded lens tissue in a hemostat. Ask an instructor if you need help. | Handle lenses only by the edges. If a lens is dirty, first remove grit with a blast of clean air or CO2. Clean the lens by wiping with a folded piece of lens paper wetted with a drop of methanol. (Do not touch the part of the tissue you use for cleaning with your fingers.) In some cases, it may be helpful to hold the folded lens tissue in a hemostat. Ask an instructor if you need help. | ||
| − | + | ===Objective lenses=== | |
Please see the Nikon [http://www.microscopyu.com/articles/optics/objectiveintro.html Introduction to Microscope Objectives] at their excellent [http://www.microscopyu.com/index.html MicroscopyU] website. | Please see the Nikon [http://www.microscopyu.com/articles/optics/objectiveintro.html Introduction to Microscope Objectives] at their excellent [http://www.microscopyu.com/index.html MicroscopyU] website. | ||
| Line 78: | Line 75: | ||
tact with the slide cover glass. After use, clean off excess oil by wicking it away with lens paper. | tact with the slide cover glass. After use, clean off excess oil by wicking it away with lens paper. | ||
| − | + | ===Sample stage=== | |
Precision [http://www.newport.com/562-Series-ULTRAlign-Precision-Multi-Axis-Positio/140089/1033/catalog.aspx Newport X/Y/Z stages] mounted on a post are available at each lab station. The stage setup is top-heavy. Avoid accidents by ensuring that the post base is always attached to an optical breadboard or table. Return the stage to the lab station when you are done with it. | Precision [http://www.newport.com/562-Series-ULTRAlign-Precision-Multi-Axis-Positio/140089/1033/catalog.aspx Newport X/Y/Z stages] mounted on a post are available at each lab station. The stage setup is top-heavy. Avoid accidents by ensuring that the post base is always attached to an optical breadboard or table. Return the stage to the lab station when you are done with it. | ||
The z-axis adjustment of the sample stage provides fine focusing. | The z-axis adjustment of the sample stage provides fine focusing. | ||
| − | + | ===Fluorescence illumination=== | |
The fluorescent illumination source is a 5 mW, λ=532 nm green laser pointer. Do not begin working with the laseruntil you are familiar with laser safety procedures. If you missed laser safety training, please see an instructor. | The fluorescent illumination source is a 5 mW, λ=532 nm green laser pointer. Do not begin working with the laseruntil you are familiar with laser safety procedures. If you missed laser safety training, please see an instructor. | ||
| − | *Wear the | + | *Wear the correct laser safety eyewear at all times — even when your laser is not on — since other groups may be using theirs. |
*Make sure the laser warning light outside the main door to the lab is flashing before you turn on a laser. | *Make sure the laser warning light outside the main door to the lab is flashing before you turn on a laser. | ||
*Never point the laser toward other people. | *Never point the laser toward other people. | ||
*Laser light can emerge from the top of the objective lens. '''Never put your face directly above the objective.''' | *Laser light can emerge from the top of the objective lens. '''Never put your face directly above the objective.''' | ||
| − | + | ===Beam expansion=== | |
| − | + | ||
Collimated light comes out of the laser pointer. The light should also be collimated when it reaches the sample (Kohler illumination). In order to provide a good image, the laser light should illuminate most of the field of view. The beam emerging from the laser pointer (abuot 1.1 mm) may not be wide enough. This means that the laser beam will need to be expanded. What expansion factor should you use for a 40× objective? You may want to be a bit conservative with beam expansion, | Collimated light comes out of the laser pointer. The light should also be collimated when it reaches the sample (Kohler illumination). In order to provide a good image, the laser light should illuminate most of the field of view. The beam emerging from the laser pointer (abuot 1.1 mm) may not be wide enough. This means that the laser beam will need to be expanded. What expansion factor should you use for a 40× objective? You may want to be a bit conservative with beam expansion, | ||
since the laser pointer is not very powerful. Overexpansion will decrease the light intensity at the sample and may not give you enough power for imaging. | since the laser pointer is not very powerful. Overexpansion will decrease the light intensity at the sample and may not give you enough power for imaging. | ||
| − | + | ===Dichroic mirror and barrier filter=== | |
| − | + | ||
The fluorophores we will emit light in the orange-red region of the visible spectrum (550-600 nm) when excited by the 532 nm green laser. | The fluorophores we will emit light in the orange-red region of the visible spectrum (550-600 nm) when excited by the 532 nm green laser. | ||
| Line 106: | Line 101: | ||
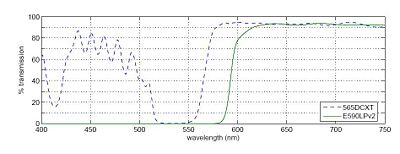
[[Image:20.309Hw3Imagespectra.JPG|center|thumb|400px|Figure 3: The transmission spectra for the 565DCXT dichroic and the E590LPv2 barrier filter]] | [[Image:20.309Hw3Imagespectra.JPG|center|thumb|400px|Figure 3: The transmission spectra for the 565DCXT dichroic and the E590LPv2 barrier filter]] | ||
| − | + | ===CCD camera=== | |
The microscope you will build does not have an eyepiece for direct visual observation. Instead, images will be captured with a Firewire-enabled CCD camera ([http://www.theimagingsource.com/en/products/cameras/firewire_mono/dmk21f04/overview/ DMK 21F04] from [http://www.theimagingsource.com/en/products/ The Imaging Source]). Its monochrome (black and white) sensor contains a grid of 640×480 square pixels that measure 5.6 μm on a side. An adapter ring converts the C-mount thread on the camera to SM1. | The microscope you will build does not have an eyepiece for direct visual observation. Instead, images will be captured with a Firewire-enabled CCD camera ([http://www.theimagingsource.com/en/products/cameras/firewire_mono/dmk21f04/overview/ DMK 21F04] from [http://www.theimagingsource.com/en/products/ The Imaging Source]). Its monochrome (black and white) sensor contains a grid of 640×480 square pixels that measure 5.6 μm on a side. An adapter ring converts the C-mount thread on the camera to SM1. | ||
| Line 112: | Line 107: | ||
==Microscope design== | ==Microscope design== | ||
| − | A basic microscope is essentially a 4-f system (sketched in Fig. 4). Some elements must be positioned precise distances apart; other distances are not critical. Use ray-tracing to determine when this is the case. | + | A basic microscope is essentially a 4-f system (sketched in Fig. 4). |
| + | |||
| + | An example microscope made by the instructors will be available in the lab for you to examine. Feel free to make improvements on this design. Stability will be crucial for the particle tracking experiments. The required specification will be achieved through good design and careful construction — not by mindlessly overtightening screws. | ||
| + | |||
| + | Some elements must be positioned precise distances apart; other distances are not critical. Use ray-tracing to determine when this is the case. | ||
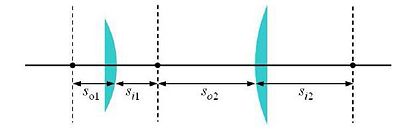
[[Image:20.309Hw3Image4flens.JPG|center|thumb|400px|Figure 4: A 4-f lens system using lenses with focal lengths f1 and f2. The object and image distances (so and si, respectively) for the lenses are indicated.]] | [[Image:20.309Hw3Image4flens.JPG|center|thumb|400px|Figure 4: A 4-f lens system using lenses with focal lengths f1 and f2. The object and image distances (so and si, respectively) for the lenses are indicated.]] | ||
| Line 136: | Line 135: | ||
* Verify your design with one of the lab instructors before beginning construction. | * Verify your design with one of the lab instructors before beginning construction. | ||
| + | ==Microscope construction and characterization== | ||
| + | The microscope should be built in two stages. First, build a white-light inverted microscope. Verify its alignment and magnification. | ||
| − | + | Next, add the fluorescence branch. An adjustable iris aperture and piece of lens or tissue paper can be very helpful for aligning the laser and directing it along the axis of the tubes. | |
| − | + | ||
| − | == | + | ===Bright field calibration=== |
| + | Make images the following samples using the three different objectives (10×, 40×, and 100×): | ||
| − | + | * The smallest line pair on the Air Force imaging target | |
| + | * A slide of 4 μm latex spheres | ||
| + | * Ronchi ruling - a periodic pattern containing 600 line-pairs per mm | ||
| − | + | Can you see all these samples? What is the magnification of the microscope and the size of its field of view? Is it what you expected? | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | Can you see all these samples? What is the magnification of the microscope and the size of its | + | |
| − | field of view? Is it what you expected? | + | |
| − | + | ||
| − | + | ||
| + | ===Fluorescence characterization and imaging=== | ||
You have the following samples available for imaging using both 40× and 100× objectives: | You have the following samples available for imaging using both 40× and 100× objectives: | ||
| − | + | *A solution slide of Rhodamine 6G (Rh6G) solution, used to adjust the microscope for uniform | |
illumination (this has its excitation peak at 530nm, and a fairly broad band of emission above | illumination (this has its excitation peak at 530nm, and a fairly broad band of emission above | ||
550nm). | 550nm). | ||
| − | + | *A sample slide with 4 μm red-fluorescent beads (modified with Nile Red dye, peak excitation | |
at 535nm, peak emission at 575nm). | at 535nm, peak emission at 575nm). | ||
Imaging tasks: | Imaging tasks: | ||
| − | + | #Use the Rh6G slide to optimize the uniformity of the illumination field (you want the light distribution hitting the sample to be as uniform as possible). Take an image of this. Use the histogram display of the camera software to be certain that the image is not overexposed. | |
| − | distribution hitting the sample to be as uniform as possible). Take an image of this. | + | #Measure the signal level while imaging Rh6G - try to achieve maximal uniformity and |
| − | + | ||
| − | + | ||
brightness. | brightness. | ||
| − | + | #Image the red beads slide. Perform flat field correction on the beads (i.e. divide the bead | |
| − | + | ||
image by the normalized Rh6G image). Compare what you see before and after flat field correction. | image by the normalized Rh6G image). Compare what you see before and after flat field correction. | ||
| − | + | If the images are noticeably different, especially at 40× (i.e. there is significant non-uniformity in the illumination field), you should improve the scope's alignment until there isn't much difference. | |
| − | ==== | + | ==Experiment 2: Particle tracking== |
| − | + | ====Introduction and background<ref>taken from http://labs.physics.berkeley.edu/mediawiki/index.php/Brownian_Motion_in_Cells</ref>==== | |
| − | + | If you have ever looked at an aqueous sample through a microscope, you have probably noticed that every small particle you see wiggles about continuously. Robert Brown, a British botanist, was not the first person to observe these motions, but perhaps the first person to recognize the significance of this observation. Experiments quickly established the basic features of these movements. Among other things, the magnitude of the fluctuations depended on the size of the particle, and there was no difference between "live" objects, such as plant pollen, and things such as rock dust. Apparently, finely crushed pieces of an Egyptian mummy also displayed these fluctuations. | |
| − | + | ||
| − | + | ||
| − | + | Brown noted: [The movements] arose neither from currents in the fluid, nor from its gradual evaporation, but belonged to the particle itself. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | This effect may have remained a curiosity had it not been for A. Einstein and M. Smoluchowski. They realized that these particle movements made perfect sense in the context of the then developing kinetic theory of fluids. If matter is composed of atoms that collide frequently with other atoms, they reasoned, then even relatively large objects such as pollen grains would exhibit random movements. This last sentence contains the ingredients for several Nobel prizes! | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | Indeed, Einstein's interpretation of Brownian motion as the outcome of continuous bombardment by atoms immediately suggested a direct test of the atomic theory of matter. J. Perrin received the 1926 Nobel Prize for validating Einstein's predictions, thus confirming the atomic theory of matter. | |
| − | + | ||
| − | + | Since then, the field has exploded, and a thorough understanding of Brownian motion is essential for everything from polymer physics to biophysics, aerodynamics, and statistical mechanics. One of the aims of this lab is to directly reproduce the experiments of J. Perrin that lead to his Nobel Prize. A translation of the key work is included in the reprints folder. Have a look – he used latex spheres, and we will use polystyrene spheres, but otherwise the experiments will be identical. In addition to reproducing Perrin's results, you will probe further by looking at the effect of varying solvent molecule size. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | {| | + | ==Experiment 3: Transport in a plant cell<ref>taken from http://labs.physics.berkeley.edu/mediawiki/index.php/Brownian_Motion_in_Cells</ref>== |
| − | |[[Image: | + | {|align="left" |
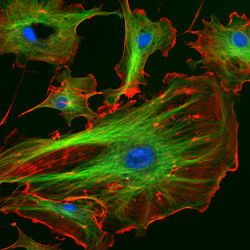
| − | | | + | | [[Image:BMC_Cytoskeleton.jpg|thumb|250px|center|The eukaryotic cytoskeleton. Actin filaments are shown in red, microtubules in green, and the nuclei are in blue.]] |
| − | | | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
|} | |} | ||
| − | + | The third part of this experiment consists of observing the motion of particles inside a living cell. Cells transport food, waste, information, etc. in membrane-bound vesicles, which are visible under a light microscope. An old-fashioned view of a cell was that it is a "bag of water" containing various enzymes in which matter is transported passively by diffusion. Though diffusion is an important mechanism, it is too slow and random for long distance transport and directing materials where they are most needed, especially in larger cells. It is now understood that cells have highly developed and intricate mechanisms for directed transport of materials. | |
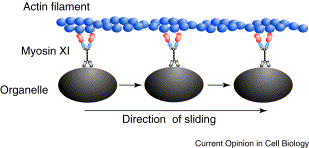
| − | + | Most motions within and of cells involve two components, a cytoskeletal fiber that serves as a track, and a [http://en.wikipedia.org/wiki/Motor_protein motor protein] that does the work. The motor molecule uses energy from the hydrolysis of one ATP molecule to bind to the fiber, bend to pull itself along the fiber, and release, all of which constitutes one "step". For an animation of this stepping process, see this [http://valelab.ucsf.edu/images/movies/mov-procmotconvkinrev5.mov movie animation] from the Vale lab web site at UC San Francisco. One can divide cellular motility mechanisms into two classes based on the cytoskeletal fibers involved. Microtubule-based mechanisms involve dynein or kinesin motors pulling on microtubules made of the protein tubulin. Actin-based mechanisms involve myosin motors pulling on actin fibers, also called microfibers. | |
| − | + | ||
| − | to | + | |
| − | to | + | |
| − | + | ||
| − | + | ||
| − | + | {| align="center" | |
| − | + | | [[Image:Myosin.gif|thumb|309px|Cartoon of myosin motors pulling organelles along an actin filament.]]|| [[Image:Kinesin.jpg|thumb|196px|Binding of kinesin motor to microtubule.]] | |
| − | + | |} | |
| − | + | ||
| − | + | ||
| − | + | Virtually all cell types exhibit directed intracellular transport, but some cell types are particularly suitable for transport studies. Fish-scale pigment cells work well, since a large fraction of the cargoes that are transported are pigmented and thus easy to observe – the disadvantage is that you would need to bring a living fish into lab as a source of these cells. For convenience, we will use epidermal cells from onion bulbs that you can easily acquire in a grocery store. With some care, a single layer of cells can be peeled off an inner section of the onion bulb and mounted flat on a slide. | |
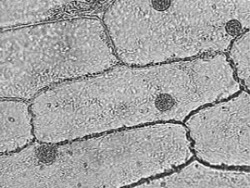
| + | {| align="center" | ||
| + | | [[Image:image005.png|thumb|250px|Onion cells in bright-field illumination. Round object in each cell is the nucleus.]] || [[Image:image003.png|thumb|250px|Vesicles in the cytoplasm of a plant cell, as seen in dark-field.]] | ||
| + | |} | ||
| − | + | In this experiment, we will be viewing the movement of vesicles within the cytoplasm of onion epidermal cells, shown above as they appear in bright-field and dark-field microscopy. The layers you see in an onion bulb develop into leaves when it sprouts. Both sides of the leaf are covered with an epidermis consisting of brick-shaped cells, each with a cell wall and cell membrane on the outside. Most of the interior of the cell is filled with a clear [http://en.wikipedia.org/wiki/Vacuole vacuole] that functions in storage and in maintenance of hydrostatic pressure essential to the stiffness of the plant (the difference between crisp lettuce and wilted lettuce). The [http://en.wikipedia.org/wiki/Cytoplasm cytoplasm], containing all of the other cell contents, occurs in a thin layer between the cell membrane and the vacuole, and in thin extensions through the vacuole called transvacuolar strands. It is within the cytoplasm that you will be observing directed transport of vesicles by an actin-based mechanism. These vesicles are spherical or rod-shaped organelles such as mitochondria, spherosomes, and [http://en.wikipedia.org/wiki/Peroxisome peroxisomes] ranging in size from 0.5 to 3 microns. The diagram of an onion cell below shows the location of the cell wall, cytoplasm and vesicles in a typical cell; you will not be able to see much of the endoplasmic reticulum or the vacuole depicted because of their transparency. Under the microscope, you will notice the vesicles are located just along the edges of the cell, or near the top and bottom surface if you focus up and down. When you see a narrow band of moving vesicles in the center of the cell, it is located in a transvacuolar strand, which may be a handy place to study motion. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | is | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | of the cell | + | |
| − | + | ||
| − | + | ||
| − | you | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
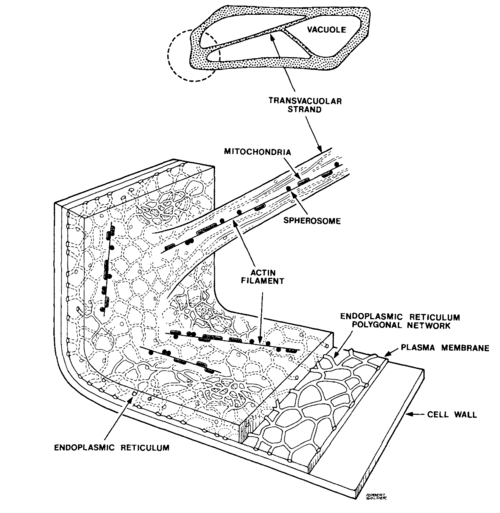
| − | + | [[Image:BMC_OnionStucture.gif|thumb|500 px|center|A 3D cross-section model of an onion epidermal cell, showing actin filaments and vesicles in the narrow bands of cytoplasm within the cell. ]] | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | In plant cells, vesicles are transported along actin fibers by myosin motor molecules. An actin filament is composed of two intertwined actin chains, about 7 nm in diameter. An actin fiber is considered structurally polar, having a (+) end and a (-) end, and most myosin motors move only towards the (+) end of the actin fiber. In order to reverse the direction of a vesicle's motion, the vesicle must grab on to another actin fiber oriented in the opposite direction. There are at least eighteen described classes of myosin, of which three, myosin VIII, XI, and XII are found in plant cells. Some myosin motors are processive, meaning that they remain bound to an actin fiber as they move step-by-step along it (analagous to this [http://valelab.ucsf.edu/images/movies/mov-procmotconvkinrev5.mov movie animation of kinesin]. Other myosins are non-processive, releasing from the actin fiber after each step. Myosin II found in muscle cells is non-processive, as illustrated in this [http://www.banyantree.org/jsale/actinmyosin/index.html video animation]. In the muscle functional unit, there are many myosin motors acting together, so there are always some attached to the actin fiber. The myosin XI responsible for transport of plant cell vesicles is the fastest myosin known and is processive. It apparently is not known how many myosin molecules are attached to the surface of a vesicle or how many of those are active at one time in pulling the vesicle along an actin fiber. | |
| − | + | ||
| − | + | ||
| − | + | In some plant cells and algal cells, a large-scale streaming motion of the cytoplasm is observed, logically called [http://en.wikipedia.org/wiki/Cytoplasmic_streaming cytoplasmic streaming]. This bulk flow is believed to be caused by myosin motors pulling the extensive endoplasmic reticulum along actin fibers lining the cell membrane. Many other vesicles are then dragged along with the endoplasmic reticulum. [http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=mcb.figgrp.5242 Lodish and Berk, et al.] provide a detailed explanation of this process and a video of cytoplasmic streaming in the pond eeed Elodea can be viewed [http://www.microscopy-uk.org.uk/mag/imgnov00/cycloa3i.avi here]. | |
| − | in the | + | |
| − | + | In your observations of vesicles in onion epidermal cells, you should distinguish between the random Brownian motion of vesicles that are unattached (or at least not actively moving along) actin filaments, the directed transport of vesicles by attached myosin motors, and possibly (though we are not sure this really happens in onions) bulk flow of vesicles in cytoplasmic streaming. | |
| − | + | ||
| − | + | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
====5.2 Experimental details==== | ====5.2 Experimental details==== | ||
| Line 370: | Line 247: | ||
Cells were cultured at 37±C in 5% CO2 in standard 100 mm £ 20 mm cell culture dishes | Cells were cultured at 37±C in 5% CO2 in standard 100 mm £ 20 mm cell culture dishes | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | ==Report Requirements== | ||
====7.1 Microscope construction==== | ====7.1 Microscope construction==== | ||
Make a sketch of your full microscope setup (a hand drawing is perfectly acceptable, but please keep | Make a sketch of your full microscope setup (a hand drawing is perfectly acceptable, but please keep | ||
| Line 509: | Line 288: | ||
{ simply outline the question you'd like to answer and suggest an approach/method/technique that | { simply outline the question you'd like to answer and suggest an approach/method/technique that | ||
you could use to test it. | you could use to test it. | ||
| + | |||
| + | <References/> | ||
</div> | </div> | ||
Revision as of 03:36, 27 October 2008
Contents
Introduction
In this lab, you will design and build a light microscope from optical components. Your instrument will be capable of two kinds of imaging: bright field transmitted light and fluorescence. In bright field,
Bright field transmitted microscopy is perhaps the simplest and most common optical microscopy method. In this technique, photons from an illuminator pass through the sample, where the may be absorbed, diffracted, or refracted. (The sample us usually mounted on a glass slide.) An objective lens on the opposite side of the sample collects the light. Most modern objective lenses produce collimated light, which is focused by a tube lens to form an image.
Illumination for fluorescence microscopy normally reaches the sample through the objective lens — from the same side of the sample that is observed. Fluorescence microscopy is normally used on samples that have been labled with a fluorescent molecule called a fluorophore. The (narrowband) illumination wavelength must match the absorption characteristic of the fluorophore. After becoming excited by a photon from the illuminator, the fluorescent molecule will emit a photon of longer wavelength. A dichroic mirror in the microscope reflects the illumination wavelength but allows the emitted photons to pass through.
The microscope design in thie lab is very flexible. Other contrast modes, such as dark field illumination and confocal, can be added to the microscope.
Objectives
- Learn about the theory and practice of light microscopy
- Use ray tracing rules to design a transmitted bright field and fluorescent light microscope
- Construct the microscope from optical components
- Characterize the microscope's performance
- Enhance and analyze microscope images
- Quantitatively track moving particles in the field of view
Roadmap and Milestones
Week 1
- Design and Build a microscope
- Characterize the transmitted bright field performance of the microscope
- Add a laser illumination beam path
Week 2
- Characterize the fluorescent imaging performance of the microscope
- Make fluorescent images
- Correct for nonuniform illumination
- Track microspheres suspended in a solvent
- Estimated diffusion coefficients
- Track the motion of vesicles under transport in a plant cell
Microscopy lab etiquette
- Observe all laser safety guidelines.
- Keep all of the boxes for the optics you use with your instrument to simplify putting things away.
- The stages are very expensive. To prevent accidents, ensure that there is a srew holding the post base to an optical breadboard or table at all times.
- There are not enough stages to go around. Remove the stage from your microscope and leave it at the lab station when you are done.
- Leave the illuminator at the lab station when you are done.
- Return objective lenses to the drawer when you are not using them.
- Never use an SM1T2 coupler without a locking ring — they are very difficult to remove if they are tightened against a lens tube or tube ring.
- Use tube rings (never an SM1T2) to mount an optic in a lens tube.
Microscope components
Rigid optical construction
Your design can use a combination to cage and lens tube components from ThorLabs. (See the ThorLabs online catalog for more details. Print catalogs are available in the lab.) Be sure you understand how to use cage cubes (C4W), cube optic mounts (B5C), and kinematic mounting plates (B4C). Please ask about any components you are not sure how to use.
An example microscope put together by the instructors will be available in the lab for you to look at. Feel free to make improvements on this design. Stability will be crucial for the particle tracking experiments. This will be achieved through good design and careful construction — not by mindlessly overtightening screws.
Simple lenses
Plano-convex spherical lenses are available with focal lengths of 25, 50, 75, 100, 125, and 200 mm. It is best to mount most optics in short (0.5" or 0.3") lens tubes. It is acceptable to mount a lens between the end of a tube and a tube ring or between two tube rings. In most cases, the convex side of the lens faces toward the collimated beam; the planar side goes toward the convergent rays. Plano-concave lenses with focal lengths of -35 and -50 are also available.
- Advice: Some students in the past have had difficulty with the Three of These Things game. Verify all optics before you use them by measuring the focal length with a ruler.
As you install lenses into your microscope, put a piece of tape on the lens tube showing focal length and orientation. This will help you both during constructino and put-away. Save the lens storage boxes and return components to the correct boxes when you are done.
Handle lenses only by the edges. If a lens is dirty, first remove grit with a blast of clean air or CO2. Clean the lens by wiping with a folded piece of lens paper wetted with a drop of methanol. (Do not touch the part of the tissue you use for cleaning with your fingers.) In some cases, it may be helpful to hold the folded lens tissue in a hemostat. Ask an instructor if you need help.
Objective lenses
Please see the Nikon Introduction to Microscope Objectives at their excellent MicroscopyU website.
There are three objective lenses available in the lab: a 10×, a 40×, and a 100×. All of these are designed for a 200 mm tube lens. An adapter ring converts the objective mounting threads to the SM1 threads used by the lens tube system.
- The back focal plane (BFP) of the objective coincides with the rear of the objective housing. This is equivalent to the focal plane of a simple lens.
- Working distance (WD) is the distance between the front end of the objective and the sample plane (when
the sample is in focus). Generally, the higher the magnification, the lower the working distance.
- The 100× objective is designed to be used with immersion oil, which provides an optical medium of pre-
determined refractive index (n = 1.5). When using the 100× objective, place a drop of oil on it. Bring the drop in con- tact with the slide cover glass. After use, clean off excess oil by wicking it away with lens paper.
Sample stage
Precision Newport X/Y/Z stages mounted on a post are available at each lab station. The stage setup is top-heavy. Avoid accidents by ensuring that the post base is always attached to an optical breadboard or table. Return the stage to the lab station when you are done with it.
The z-axis adjustment of the sample stage provides fine focusing.
Fluorescence illumination
The fluorescent illumination source is a 5 mW, λ=532 nm green laser pointer. Do not begin working with the laseruntil you are familiar with laser safety procedures. If you missed laser safety training, please see an instructor.
- Wear the correct laser safety eyewear at all times — even when your laser is not on — since other groups may be using theirs.
- Make sure the laser warning light outside the main door to the lab is flashing before you turn on a laser.
- Never point the laser toward other people.
- Laser light can emerge from the top of the objective lens. Never put your face directly above the objective.
Beam expansion
Collimated light comes out of the laser pointer. The light should also be collimated when it reaches the sample (Kohler illumination). In order to provide a good image, the laser light should illuminate most of the field of view. The beam emerging from the laser pointer (abuot 1.1 mm) may not be wide enough. This means that the laser beam will need to be expanded. What expansion factor should you use for a 40× objective? You may want to be a bit conservative with beam expansion, since the laser pointer is not very powerful. Overexpansion will decrease the light intensity at the sample and may not give you enough power for imaging.
Dichroic mirror and barrier filter
The fluorophores we will emit light in the orange-red region of the visible spectrum (550-600 nm) when excited by the 532 nm green laser.
In any fluorescence system, a key concern is viewing only the emitted fluorescence photons, and eliminating any background light, especially from the illumination source. Two optical elements address the problem. A dichroic mirror passes light of one wavelength, and reflects light of another. The transmission spectrum for the 565DCXT dichroicis shown in Fig. 3. A barrier filter blocks a particular spectral region. We will use the E590LPv2 barrier filter.
Figure 1 shows the tranmittance of the dichroic mirror and barrier filter over a range of wavelengths. The barrier filter is essential for high-sensitivity fluorescence imaging. It will pass the red light from the LED in the illuminator for bright field imaging as well as the fluorescence photons.
CCD camera
The microscope you will build does not have an eyepiece for direct visual observation. Instead, images will be captured with a Firewire-enabled CCD camera (DMK 21F04 from The Imaging Source). Its monochrome (black and white) sensor contains a grid of 640×480 square pixels that measure 5.6 μm on a side. An adapter ring converts the C-mount thread on the camera to SM1.
The barrier filter, SM1 adapter, and a quick-connect have been installed on the camera. Please leave the camera at the lab station when you are done with it. Camera software called IC Capture is installed on the labstation PCs.
Microscope design
A basic microscope is essentially a 4-f system (sketched in Fig. 4).
An example microscope made by the instructors will be available in the lab for you to examine. Feel free to make improvements on this design. Stability will be crucial for the particle tracking experiments. The required specification will be achieved through good design and careful construction — not by mindlessly overtightening screws.
Some elements must be positioned precise distances apart; other distances are not critical. Use ray-tracing to determine when this is the case.
Bright field transmitted illumination imaging
Sketch out a rough design for your microscope on paper. Begin with the bright field illumination path.
Note:
- The Nikon objective lenses are designed to be paired with a 200 mm tube lens.
- Assume that the objectives behave as ideal plano-convex lenses.
- Fine focusing will be achieved by adjusting the height of the sample stage.
- Start the alignment with a 10× objective but progress to 40× and 100×.
- Use the LED illuminators for bright field transmitted light imaging.
- Put a quick connect in your design such that the camera CCD will end up 200mm from the back focal plane of the objective. Remember that the CCD is recessed inside the opening of the camers.
- The barrier filter has been installed in a lens tube already attached to the camera.
Fluorescence imaging
Now sketch a layout that includes both the bright field and fluorescence illumination paths.
- Use a dichroic to direct the laser illumination toward the sample and to pass the emitted (fluorescent) light back through to the CCD.
- The barrier filter removes any light from the illumination laser that was not reflected by the dichroic.
- During fluorescent imaging, you will not use the bright field illuminator. Bright field capability is useful for first visualizing the sample and viewing what features are in the field of view. Your design should retain the ability to do both white light and fluorescent visualization.
- Verify your design with one of the lab instructors before beginning construction.
Microscope construction and characterization
The microscope should be built in two stages. First, build a white-light inverted microscope. Verify its alignment and magnification.
Next, add the fluorescence branch. An adjustable iris aperture and piece of lens or tissue paper can be very helpful for aligning the laser and directing it along the axis of the tubes.
Bright field calibration
Make images the following samples using the three different objectives (10×, 40×, and 100×):
- The smallest line pair on the Air Force imaging target
- A slide of 4 μm latex spheres
- Ronchi ruling - a periodic pattern containing 600 line-pairs per mm
Can you see all these samples? What is the magnification of the microscope and the size of its field of view? Is it what you expected?
Fluorescence characterization and imaging
You have the following samples available for imaging using both 40× and 100× objectives:
- A solution slide of Rhodamine 6G (Rh6G) solution, used to adjust the microscope for uniform
illumination (this has its excitation peak at 530nm, and a fairly broad band of emission above 550nm).
- A sample slide with 4 μm red-fluorescent beads (modified with Nile Red dye, peak excitation
at 535nm, peak emission at 575nm).
Imaging tasks:
- Use the Rh6G slide to optimize the uniformity of the illumination field (you want the light distribution hitting the sample to be as uniform as possible). Take an image of this. Use the histogram display of the camera software to be certain that the image is not overexposed.
- Measure the signal level while imaging Rh6G - try to achieve maximal uniformity and
brightness.
- Image the red beads slide. Perform flat field correction on the beads (i.e. divide the bead
image by the normalized Rh6G image). Compare what you see before and after flat field correction.
If the images are noticeably different, especially at 40× (i.e. there is significant non-uniformity in the illumination field), you should improve the scope's alignment until there isn't much difference.
Experiment 2: Particle tracking
Introduction and background[1]
If you have ever looked at an aqueous sample through a microscope, you have probably noticed that every small particle you see wiggles about continuously. Robert Brown, a British botanist, was not the first person to observe these motions, but perhaps the first person to recognize the significance of this observation. Experiments quickly established the basic features of these movements. Among other things, the magnitude of the fluctuations depended on the size of the particle, and there was no difference between "live" objects, such as plant pollen, and things such as rock dust. Apparently, finely crushed pieces of an Egyptian mummy also displayed these fluctuations.
Brown noted: [The movements] arose neither from currents in the fluid, nor from its gradual evaporation, but belonged to the particle itself.
This effect may have remained a curiosity had it not been for A. Einstein and M. Smoluchowski. They realized that these particle movements made perfect sense in the context of the then developing kinetic theory of fluids. If matter is composed of atoms that collide frequently with other atoms, they reasoned, then even relatively large objects such as pollen grains would exhibit random movements. This last sentence contains the ingredients for several Nobel prizes!
Indeed, Einstein's interpretation of Brownian motion as the outcome of continuous bombardment by atoms immediately suggested a direct test of the atomic theory of matter. J. Perrin received the 1926 Nobel Prize for validating Einstein's predictions, thus confirming the atomic theory of matter.
Since then, the field has exploded, and a thorough understanding of Brownian motion is essential for everything from polymer physics to biophysics, aerodynamics, and statistical mechanics. One of the aims of this lab is to directly reproduce the experiments of J. Perrin that lead to his Nobel Prize. A translation of the key work is included in the reprints folder. Have a look – he used latex spheres, and we will use polystyrene spheres, but otherwise the experiments will be identical. In addition to reproducing Perrin's results, you will probe further by looking at the effect of varying solvent molecule size.
Experiment 3: Transport in a plant cell[2]
The third part of this experiment consists of observing the motion of particles inside a living cell. Cells transport food, waste, information, etc. in membrane-bound vesicles, which are visible under a light microscope. An old-fashioned view of a cell was that it is a "bag of water" containing various enzymes in which matter is transported passively by diffusion. Though diffusion is an important mechanism, it is too slow and random for long distance transport and directing materials where they are most needed, especially in larger cells. It is now understood that cells have highly developed and intricate mechanisms for directed transport of materials.
Most motions within and of cells involve two components, a cytoskeletal fiber that serves as a track, and a motor protein that does the work. The motor molecule uses energy from the hydrolysis of one ATP molecule to bind to the fiber, bend to pull itself along the fiber, and release, all of which constitutes one "step". For an animation of this stepping process, see this movie animation from the Vale lab web site at UC San Francisco. One can divide cellular motility mechanisms into two classes based on the cytoskeletal fibers involved. Microtubule-based mechanisms involve dynein or kinesin motors pulling on microtubules made of the protein tubulin. Actin-based mechanisms involve myosin motors pulling on actin fibers, also called microfibers.
Virtually all cell types exhibit directed intracellular transport, but some cell types are particularly suitable for transport studies. Fish-scale pigment cells work well, since a large fraction of the cargoes that are transported are pigmented and thus easy to observe – the disadvantage is that you would need to bring a living fish into lab as a source of these cells. For convenience, we will use epidermal cells from onion bulbs that you can easily acquire in a grocery store. With some care, a single layer of cells can be peeled off an inner section of the onion bulb and mounted flat on a slide.
In this experiment, we will be viewing the movement of vesicles within the cytoplasm of onion epidermal cells, shown above as they appear in bright-field and dark-field microscopy. The layers you see in an onion bulb develop into leaves when it sprouts. Both sides of the leaf are covered with an epidermis consisting of brick-shaped cells, each with a cell wall and cell membrane on the outside. Most of the interior of the cell is filled with a clear vacuole that functions in storage and in maintenance of hydrostatic pressure essential to the stiffness of the plant (the difference between crisp lettuce and wilted lettuce). The cytoplasm, containing all of the other cell contents, occurs in a thin layer between the cell membrane and the vacuole, and in thin extensions through the vacuole called transvacuolar strands. It is within the cytoplasm that you will be observing directed transport of vesicles by an actin-based mechanism. These vesicles are spherical or rod-shaped organelles such as mitochondria, spherosomes, and peroxisomes ranging in size from 0.5 to 3 microns. The diagram of an onion cell below shows the location of the cell wall, cytoplasm and vesicles in a typical cell; you will not be able to see much of the endoplasmic reticulum or the vacuole depicted because of their transparency. Under the microscope, you will notice the vesicles are located just along the edges of the cell, or near the top and bottom surface if you focus up and down. When you see a narrow band of moving vesicles in the center of the cell, it is located in a transvacuolar strand, which may be a handy place to study motion.
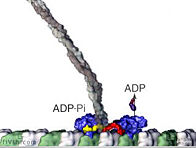
In plant cells, vesicles are transported along actin fibers by myosin motor molecules. An actin filament is composed of two intertwined actin chains, about 7 nm in diameter. An actin fiber is considered structurally polar, having a (+) end and a (-) end, and most myosin motors move only towards the (+) end of the actin fiber. In order to reverse the direction of a vesicle's motion, the vesicle must grab on to another actin fiber oriented in the opposite direction. There are at least eighteen described classes of myosin, of which three, myosin VIII, XI, and XII are found in plant cells. Some myosin motors are processive, meaning that they remain bound to an actin fiber as they move step-by-step along it (analagous to this movie animation of kinesin. Other myosins are non-processive, releasing from the actin fiber after each step. Myosin II found in muscle cells is non-processive, as illustrated in this video animation. In the muscle functional unit, there are many myosin motors acting together, so there are always some attached to the actin fiber. The myosin XI responsible for transport of plant cell vesicles is the fastest myosin known and is processive. It apparently is not known how many myosin molecules are attached to the surface of a vesicle or how many of those are active at one time in pulling the vesicle along an actin fiber.
In some plant cells and algal cells, a large-scale streaming motion of the cytoplasm is observed, logically called cytoplasmic streaming. This bulk flow is believed to be caused by myosin motors pulling the extensive endoplasmic reticulum along actin fibers lining the cell membrane. Many other vesicles are then dragged along with the endoplasmic reticulum. Lodish and Berk, et al. provide a detailed explanation of this process and a video of cytoplasmic streaming in the pond eeed Elodea can be viewed here.
In your observations of vesicles in onion epidermal cells, you should distinguish between the random Brownian motion of vesicles that are unattached (or at least not actively moving along) actin filaments, the directed transport of vesicles by attached myosin motors, and possibly (though we are not sure this really happens in onions) bulk flow of vesicles in cytoplasmic streaming.
5.2 Experimental details
5.2.1 Stability and setup
The major challenge of particle tracking microrheometery is the small scale of the thermal forces and the associated nanometer scale displacements. A few things you can do to ensure the experiment works: Be sure all the microscope components are rigidly assembled and ¯rmly tightened. Poorly built scopes shake. It is also vital that when you perform this experiment that the optical tables are °oating so that building noise is isolated. Of course, avoid touching the optical table and the microscope during the measurement. There are also cardboard boxes available that you can put over your microscope to isolate it from air currents. Finally, make sure that you and the people around you are not talking too loudly during the experiment, because acoustic noise is signi¯cant. The camera gain and brightness setting should be set as described in Figure 7.
5.2.2 System veri¯cation
To verify that your system is su±ciently rigid/stable, ¯rst measure a specimen containing 1 m red °uorescent beads (Molecular Probes) dried in a cell dish. Chose a ¯eld of view in which you can see at least 3-4 beads. Using a 40£ objective record an .avi movie for about 3 min. at a frame rate of 30 frames/sec. From your experience with image processing, you already know how to import .avi movie data into matlab. To improve signal to noise ratio, sum every 30 frames together, which will make your sequence have a temporal interval of 1 sec. Use the bead tracking processing algorithm on two beads to calculate two trajectories. To further reduce common-mode motion from room vibrations, calculate the di®erential trajectory from the individual trajectories of these two beads. Calculate the MSD h¢r2i from this di®erential trajectory. Your MSD should start out less than 10 nm2 at ¿ = 1 sec. and still be less than 100 nm2 for ¿ = 180 sec. If you don't get this, do not proceed further and ask for help. 5.2.3 Live cell measurements Now that your system is su±ciently stable, you can run the experiment on cell samples. A key technique to keep in mind when working with live cells { to avoid shocking them with \cold at 20±C, be sure that any solutions you add are pre-warmed to 37±C. We will keep a warm-water bath running on the hotplate for this purpose, in which we will keep the various media. You are provided with NIH 3T3 ibroblasts, which were prepared as follows: Cells were cultured at 37±C in 5% CO2 in standard 100 mm £ 20 mm cell culture dishes
Report Requirements
7.1 Microscope construction
Make a sketch of your full microscope setup (a hand drawing is perfectly acceptable, but please keep it neat - a ruler is handy) with important parameters indicated (i.e. lens focal lengths, distances of main components, etc.).
7.2 Experiment 1: microscope characterization and Fourier-plane imaging
1. Include your favorite 2-3 white-light images, indicating the magni¯cation and ¯eld of view (do they match what was expected?). 2. Include an uncorrected and a °at-¯eld-corrected °uorescent image of the 4 ¹m beads. 3. Calculate the di®raction-limited resolution for the 10£, 40£ and 100£ objectives, and how this compares with which samples each objective could or could not resolve. 4. Include both the real and Fourier-plane images of the peacock feather and tissue paper, and quantitatively describe how they relate to each other?
7.3 Experiment 2: microrheology measurements by particle tracking
Analyze the bead trajectories for the normal and CytoD-treated cells. Extract their MSD h¢r2i, and the G0 and G00 modulus values, and include enough detail to make it clear how you performed the analysis. Do you get consistent results across multiple cells in each group? What can you tell about the e®ect of CytoD on the mechanical properties of these cells?
7.4 Experiment 3: °uorescence imaging of the actin cytoskeleton
Include one or two of your favorite actin cytoskeleton images from each of the CytoD treated and untreated cell groups. Use your image processing knowledge to optimize image quality. Apply the algorithms you developed during the image processing lab to quantify the degree of \¯berness of the actin structures with and without CytoD. As in Experiment 2, what can you say about the e®ect of CytoD on these cells? Discuss whether the results of this experiment and the microrheology experiment are consistent with each other?
Bonus (optional)
You've now used two di®erent optical microscopy methods to study the e®ects of cytochalasin D on NIH 3T3 ¯broblasts. Hopefully, as you were working, many questions arose in your mind about di®erent cell properties, their underlying physics, experimental conditions, etc. For bonus credit, think about and propose any other experiments you might like to do (using these microscopes, or the AFMs, or any other approach you like) to study any related questions of cytoskeletal mechanics, microrheology, °uorescent labeling, etc. This is not a formal grant proposal { simply outline the question you'd like to answer and suggest an approach/method/technique that you could use to test it.