DNA Melting: Simulating DNA Melting - Intermediate Topics
Contents
Introduction
This tutorial will show you how to simulate the DNA melting experiment in Matlab. It will be essential to have your data analysis scripts working before you begin running experiments in the lab. You can use a simulated dataset generated by the code in this tutorial to help debug your data analysis scripts.
Code from this page can be copied into a Matlab command window. M-files are available in the course locker.
If you prefer to work in Python, please see this page.
Before you begin
- Read the DNA Melting Lab Manual.
- Review the DNA Melting Thermodynamics Lecture Notes.
- If you need a review of Matlab fundamentals, see Matlab Fundamentals tutorial.
Procedure
One of the goals of this lab is to estimate $ \left . T_m \right . $, $ \Delta H^{\circ} $, and $ \Delta S^{\circ} $ for the annealing of several DNA samples. All three of these quantities will be estimated from a melting curve (a plot of the fraction of dsDNA versus temperature) and its derivative.
During each experimental run, the range of temperatures will be provided by natural cooling of the sample after it has been heated in a bath. Temperature will be derived from the voltage across an RTD affixed to the heating block. DsDNA concentration will be estimated from the fluorescence intensity of the sample. Your data analysis script will have to convert the RTD and fluorescence intensity voltage signals into estimates of the temperature and dsDNA fraction.
Since both voltages depend on sample temperature, the first step will be to simulate natural cooling of the sample. From there, RTD and fluorescence voltages can be derived.
You will certainly notice some noise on the signals you capture. (See: Where does electonic noise come from?.) Noise will be an extremely important (and annoying) factor in this experiment because the melting curve will be differentiated. Differentiation emphasizes noise. (Think about the frequency response of a differentiator.) In order to ensure that your data analysis script effectively handles the noise you are likely to observe, simulated random and impulse noise will be added to the signals.
Simulate cooling of the sample
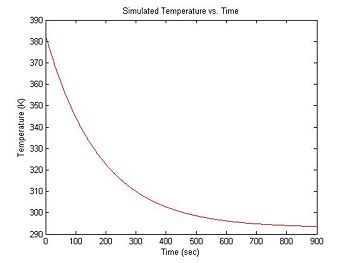
At the beginning of an experimental run, the DNA + fluorescent dye sample is heated in a bath to around 90°C. Over a period of about 15 minutes, the sample cools to room temperature (about 20°C). Cooling can be approximated by an exponential function with a time constant of 3 minutes. The equation for temperature (in °K) versus time is:
- $ T(t) = 90 e ^ {-\frac{t}{180}} + 293 $
In Matlab:
simulationLength = 900 % 15 minute simulation
sampleRate = 1 % 1 Hz sample rate
t = 0:(1./sampleRate):simulationLength; % create a time vector (units: seconds)
T = 90 * exp(-1 * t ./ 180) + 293; % exponential cooling from 383K to 293K
plot(t, T, 'r');
xlabel('Time (sec)');
ylabel('Temperature (K)');
title('Simulated Temperature vs. Time');
If you have any questions about the element-by-element division operator in Matlab (./) see:this page.
Convert sample temperature to RTD Voltage
Model the voltage divider
The RTD device does not measure temperature directly. The resistance of the RTD is $ R_{RTD} = 1000 + 3.85(T - 273) $. In the experimental apparatus, the RTD is hooked up as a voltage divider with another resistor, R. The power supply is 15 volts. Thus, the output voltage is: $ V_{RTD} = 15 * R_{RTD}/(R + R_{RTD}) $. The following code converts the tepmerature vector T into the equivalent RTD voltage measurements, assuming R = 20KΩ.
R_RTD = 1000 + 3.85 * (T - 273); % Compute RTD resistance in Ohms
V_RTD = 15 * R_RTD ./ (20E3 + R_RTD); % Compute V_RTD in Volts
plot(t, V_RTD, 'r');
xlabel('Time (sec)');
ylabel('RTD Voltage (V)');
title('Simulated RTD Voltage vs. Time');
Add some noise
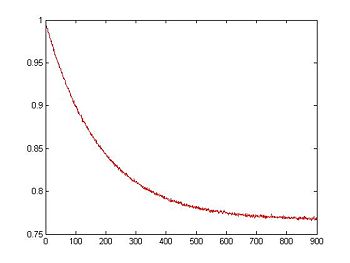
To simulate noise, we can use Matlab's randn function, which generates normally distributed random numbers. The two arguments tell randn to create a vector of random samples with size 1xlength(V_RTD).
noise = 0.001 * randn(1,length(V_RTD)); % Create noise vector, standard deviation=1mV
V_RTD = V_RTD + noise; % Add simulated noise to the voltage signal
plot(t, V_RTD, 'r');
xlabel('Time (sec)');
ylabel('RTD Voltage (V)');
title('Simulated RTD Voltage with Noise vs. Time');
Model sample fluorescence signal
Now that we have a simulated RTD voltage signal, the next step is to simulate the fluorescence signal. Remember that fluorescence intensity is proportional to the amount of dsDNA in the sample.
Write a function to compute f
The dsDNA concentration at a given temperature can be computed with the equation that we derived in class. Since this is a very complicated equation and we will use it many times, it makes sense to write a Matlab function. The file is available online here.
<include src="http://web.mit.edu/~20.309/www/Matlab/DnaFraction.m" />
Using the element-by-element divide operator makes it possible for the DnaFraction function to accept either a single value of temperature or a vector of temperature samples. (As a general rule, use the ./ operator instead of / unless you intend to do a matrix divide.)
Create a simulated fluorescence curve
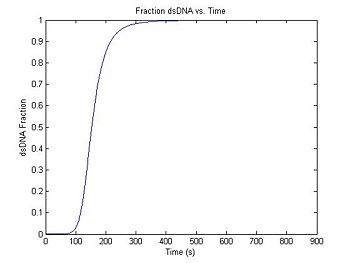
Since DnaFraction can take a vector as an argument, generating the fluorescence curve is simple.
f = DnaFraction(1E-6, T, -184, -71E3);
plot(t,f)
title('Fraction dsDNA vs. Time');
xlabel('Time (s)');
ylabel('dsDNA Fraction');
Scale the curve and add some noise
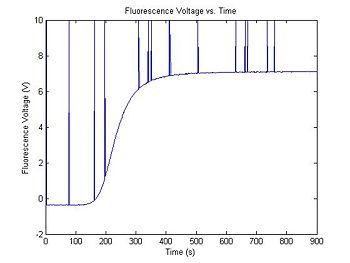
The output range of the transimpedance amplifier over the course of an experimental run can vary depending on several factors, including the amplifier gain, optical system gain, etc... A typical setup might produce values between, say, 0.5 and 7.2 Volts. The simulation code generates two random numbers to set the range and minimum value of the data. Your data analysis script will have to normalize the amplifier output voltage to a fractional value between 0 and 1. (What assumptions will you make about dsDNA concentration to do the normalization?)
In addition to Gaussian noise, the high gain amplifier can produce impulse noise. (This can be caused by small static discharges.) It is a good idea to include some filtering for impulse noise in your data analysis script.
Range = 5 + 4 * rand(); % Range between 5 and 9
Minimum = 2 * rand() - 1; % Minimum between +/- 1
V_f = Range * f + Minimum; % Scale output
noise = 0.01 * randn(1,length(V_f)); % Random noise, standard deviation=10mV
V_f = V_f + noise; % Add random noise
noise = rand(1,length(V_f)); % Uniformly distributed random numbers
ImpulseNoiseProbability = 1 ./ 75; % Probability of impulse noise
noise = 20 * (noise < ImpulseNoiseProbability) - 10; % Quantize impulse noise
V_f = max(V_f, noise); % Add in impulse noise
plot(t, V_f)
title('Fluorescence Voltage vs. Time');
xlabel('Time (s)');
ylabel('Fluorescence Voltage (V)');
Data analysis
Tm can be estimated directly from the melting curve or by finding the peak of the melting curve's derivative.
An outline of the data analysis procedure is:
- Filter V_RTD and V_f to remove noise.
- Transform RTD voltage to temperature.
- Transform photodiode current to relative fluorescence.
- Ensure that the melting function is uniquely values by combining samples with identical temperature values.
- Differentiate the resulting function with respect to temperature.