Difference between revisions of "Assignment 9, Part 1: Analyze two-color yeast images"
Juliesutton (Talk | contribs) |
Juliesutton (Talk | contribs) (→Getting Started) |
||
| Line 3: | Line 3: | ||
Start by downloading Assignment9TestData.mat. This is an example of the data you will be collecting in assignment 10 with an 8 minute valve oscillation period. The movie was collected using the same 20.309 microscope that you built in Assignment 8. During an oscillation experiment, the microscope first records an image with blue illumination followed by one with green illumination. It had a 40x objective and a 125 mm tube lens - remember that results in a 25X magnification. | Start by downloading Assignment9TestData.mat. This is an example of the data you will be collecting in assignment 10 with an 8 minute valve oscillation period. The movie was collected using the same 20.309 microscope that you built in Assignment 8. During an oscillation experiment, the microscope first records an image with blue illumination followed by one with green illumination. It had a 40x objective and a 125 mm tube lens - remember that results in a 25X magnification. | ||
| − | In MATLAB, display the contents of the structure in | + | The sample is similar to the one in Mettetal et al.: we are using ''S. Cerevisiae'' with the protein Hog1 fused to GFP (which is excited by blue light) and an mRNA binding protein (we'll call MCP) fused to RFP (which is excited by green light) to locate the nucleus. Our goal is to calculate the Hog1 intensity in the nucleus vs the cytosol as a function of the high/low valve state. |
| + | |||
| + | In MATLAB, display the contents of the structure in the data file: | ||
<pre>twoColorYeastTest = | <pre>twoColorYeastTest = | ||
| Line 24: | Line 26: | ||
{{Template:Assignment Turn In|message = Choose a single frame of the movie. Make a four panel figure and display: the blue image, the green image (both with a scale bar), and their corresponding image histograms. Include an appropriate title for each panel. }} | {{Template:Assignment Turn In|message = Choose a single frame of the movie. Make a four panel figure and display: the blue image, the green image (both with a scale bar), and their corresponding image histograms. Include an appropriate title for each panel. }} | ||
| + | |||
| + | ==Finding Cells and Nuclei== | ||
| + | |||
| + | For each image, we need to find the locations of the cells and nuclei. We'll use a familiar method of locating the nuclei using the green image by setting a threshold to find the bright regions. In addition, we'll use a morphological opening operation (an erosion followed by a dilation) to remove hot pixels or other noise. | ||
| + | |||
| + | <pre> | ||
| + | bwImage = imbinarize(NucleiFrame,NucleiThreshold); | ||
| + | nucleiMask = imopen(bwImage,strel('disk',2)); | ||
| + | </pre> | ||
| + | |||
| + | Instead of using a threshold to find the cells in the blue image, we'll use <tt>imfindcircles</tt>, which implements the Hough transform. The benefit of this method over using a threshold, is that it will allow us to identify single cells within a larger clump. Here's a typical implementation: | ||
| + | |||
| + | <pre> | ||
| + | [cellCenters, cellRadii] = imfindcircles(CellFrame,[Rmin Rmax],'ObjectPolarity','bright','Sensitivity',0.95); | ||
| + | </pre> | ||
| + | where <tt>Rmin</tt> and <tt>Rmax</tt> are the smallest and largest expected radii. | ||
| + | |||
| + | [[Image:FindingCellsAndNucleiExample.png|center|thumb|400px]] | ||
Revision as of 17:25, 12 November 2018
Getting Started
Start by downloading Assignment9TestData.mat. This is an example of the data you will be collecting in assignment 10 with an 8 minute valve oscillation period. The movie was collected using the same 20.309 microscope that you built in Assignment 8. During an oscillation experiment, the microscope first records an image with blue illumination followed by one with green illumination. It had a 40x objective and a 125 mm tube lens - remember that results in a 25X magnification.
The sample is similar to the one in Mettetal et al.: we are using S. Cerevisiae with the protein Hog1 fused to GFP (which is excited by blue light) and an mRNA binding protein (we'll call MCP) fused to RFP (which is excited by green light) to locate the nucleus. Our goal is to calculate the Hog1 intensity in the nucleus vs the cytosol as a function of the high/low valve state.
In MATLAB, display the contents of the structure in the data file:
twoColorYeastTest =
struct with fields:
Movie: [544×728×2×16 double]
Time: [16×2 double]
ValveOscillationPeriod: 480
BlueCameraGainAndExposure: [5 5000000]
GreenCameraGainAndExposure: [15 5000000]
ValveState: [16×1 logical]
The data structure contains all the important information from the experiment:
- A two-color movie, where the 3d dimension is the color (1 = Blue, 2 = Green), and the fourth is the frame number. Notice that twoColorYeastTest.Movie has already converted to a double, but it is not yet normalized by 4095.
- The time stamp of each frame recorded, in seconds since the start of the experiment. Column 1 contains the timestamps of the Blue images, column 2 is for the green images.
- The oscillation valve period, in seconds.
- The blue and green image camera settings in the format [gain, exposure], where exposure is in microseconds.
- The valve state (0 for low salt, 1 for high salt) at the time of each image. If you need to know the valve state more precisely, it can be calculated with the following conditional statement: $ sin(2 \pi t/T) \ge 0 $, where T is the valve oscillation period, and t is the time since the start of the experiment (this is how the state is determined in the software).
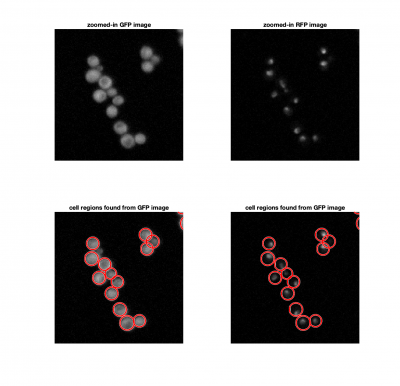
Finding Cells and Nuclei
For each image, we need to find the locations of the cells and nuclei. We'll use a familiar method of locating the nuclei using the green image by setting a threshold to find the bright regions. In addition, we'll use a morphological opening operation (an erosion followed by a dilation) to remove hot pixels or other noise.
bwImage = imbinarize(NucleiFrame,NucleiThreshold);
nucleiMask = imopen(bwImage,strel('disk',2));
Instead of using a threshold to find the cells in the blue image, we'll use imfindcircles, which implements the Hough transform. The benefit of this method over using a threshold, is that it will allow us to identify single cells within a larger clump. Here's a typical implementation:
[cellCenters, cellRadii] = imfindcircles(CellFrame,[Rmin Rmax],'ObjectPolarity','bright','Sensitivity',0.95);
where Rmin and Rmax are the smallest and largest expected radii.