Assignment 4 part 3: Track microspheres over time
This is Part 3 of Assignment 4.
Overview
In this part of the assignment, you will develop code to track fluorescent microspheres in a sequence of frames. You'll check your code with a synthetic movie of diffusing beads, before recoding actual movies of fluorescent 0.84 μm microspheres diffusing in glycerol solutions. From these movies, you'll plot the mean squared displacement and measure the diffusion coefficient - and thus the viscosity of the glycerol.
Next week, you will use the same code to characterize the diffusive motion of particles in different rheological environments (including live cells).
Develop particle tracking code
Generate a synthetic movie of diffusing microspheres using the function SimulatedBrownianMotionMovie below (you may need to make some small modifications to the code to get it to work...).
function SimulatedMovie = SimulatedBrownianMotionMovie( varargin )
% this is a fancy way to take care of input arguments
% all of the arguments have default values
% if you want to change a default value, call this function with name/value pairs like this:
% foo = SimulatedBrownianMotionMovie( 'NumerOfParticles', 10, 'ParticleDiameter' 3.2E-6 );
% it's safe to not understand this, if you like.
p = inputParser();
p.addParameter( 'NumberOfParticles', 5 );
p.addParameter( 'ParticleDiameter', 1E-6 );
p.addParameter( 'PixelSize', 7.4E-6 / 40 );
p.addParameter( 'ImageSize', [ 300 300 ] );
p.addParameter( 'FrameInterval', 1/10 );
p.addParameter( 'TemperatureKelvin', 293 );
p.addParameter( 'Viscosity', 1.0E-3 );
p.addParameter( 'BoltzmannConstant', 1.38e-23 );
p.addParameter( 'NumberOfFrames', 100 );
p.addParameter( 'ShowImages', true );
p.parse( varargin{:} );
assignLocalVariable = @( name, value ) assignin( 'caller', name, value );
allParameters = fields( p.Results );
for ii = 1:numel(allParameters)
assignLocalVariable( allParameters{ii}, p.Results.(allParameters{ii}) );
end
% at this point, there will be local scope variables with the names of
% each of the parameters specified above
close all
diffusionCoefficient = BoltzmannConstant * TemperatureKelvin / ( 3 * pi * Viscosity * ParticleDiameter );
displacementStandardDeviation = sqrt( diffusionCoefficient * 2 * FrameInterval );
% create initial particle table with random positions
particleRadiusInPixels = ParticleDiameter / 2 / PixelSize;
Position = bsxfun( @times, rand( NumberOfParticles, 2 ), ImageSize );
Radius = particleRadiusInPixels * ones( NumberOfParticles, 1 );
TotalPhotonNumber = sqrt(2) * particleRadiusInPixels^2 * 4095 * ones( NumberOfParticles, 1 );
particleTable = table( Position, Radius, TotalPhotonNumber );
SimulatedMovie = zeros( ImageSize(1), ImageSize(2), NumberOfFrames );
figure
for ii = 1:NumberOfFrames
ii
SimulatedMovie(:,:,ii) = SyntheticFluorescentMicrosphereImageWithNoise( particleTable, ImageSize ) / 4095;
if( ShowImages )
imshow( SimulatedMovie(:,:,ii) );
drawnow
end
% update the particle position according to the diffusion
oldPosition = table2array( particleTable(:, 'Position') );
Position = oldPosition + displacementStandardDeviation / PixelSize * randn( size( oldPosition ) );
% coefficient
particleTable(:, 'Position') = table( Position );
end
end
Use the code you developed in parts 1 and 2 of this assignment to write a function that finds the weighted centroids of the particles in each frame of a movie. Make sure to use the WeightedCentroid property instead of Centroid in the regionprops command (and be sure you understand the difference between the two!). Your function should return an N x 3 matrix with the following three columns (in order):
- the Y centroid coordinate,
- the X centroid coordinate, and
- the frame number.
Test out your function using your synthetic movie.
Once you have your N x 3 matrix, use the track function (an announcement with a link to this function was sent out to the class) to add a fourth column to the matrix containing a unique identifier for each particle. The N x 4 matrix will be the input to CalculateDiffusionCoefficientsFromTrajectories (below).
Diffusion measurements from particle tracking data
The function CalculateDiffusionCoefficientFromTrajectories can be used to plot the MSD as a function of time for fluorescent particles and calculate their average diffusion coefficient. The input argument Centroids can be generated by the function track, which creates a 4 column matrix listing the x- and y- coordinates, the frame number and the particle number. Additional input arguments to CalculateDiffusionCoefficientFromTrajectories include the pixel size, the movie frame rate, and an optional figure handle for plotting the MSD.
function out = CalculateDiffusionCoefficientFromTrajectories(Centroids, PixelSize, FrameRate, FigureHandle)
Centroids( :, 1:2 ) = Centroids( :, 1:2 ) * PixelSize;
numberOfParticles = max( Centroids(:,4) );
particles = cell( 1, numberOfParticles );
longestTrack = 0;
particlesToSkip = [];
if( nargin < 4 || isempty( FigureHandle ))
FigureHandle = figure;
end
figure( FigureHandle )
for ii = 1:numberOfParticles
% get the centroids for a single particle
particle = Centroids(Centroids(:,4) == ii, :);
numberOfSamples = size(particle,1); % number of samples in trajectory
particles{ii} = struct();
numberOfMsds = round(numberOfSamples-1);% / 8);
particles{ii}.msd = zeros(1, numberOfMsds);
particles{ii}.msdUncertainty = zeros(1, numberOfMsds);
particles{ii}.number = numberOfMsds;
if numberOfMsds==0
particlesToSkip = [particlesToSkip ii];
else
for tau = 1:numberOfMsds
dx = particle(1:1:(end-tau), 1) - particle((tau+1):1:end, 1);
dy = particle(1:1:(end-tau), 2) - particle((tau+1):1:end, 2);
drSquared = dx.^2+dy.^2;
msd = mean( drSquared );
particles{ii}.msd(tau) = msd;
particles{ii}.msdUncertainty(tau) = std( drSquared ) / sqrt(length( drSquared ));
end
particles{ii}.diffusionCoefficient = particles{ii}.msd(1) / (2 * 2 * FrameRate^-1);
longestTrack = max( longestTrack, numberOfSamples );
loglog( (1:numberOfMsds)/FrameRate, particles{ii}.msd(1:numberOfMsds), 'color', rand(1,3) );
hold on;
end
end
% compute the average value of D and the ensemble average of msd
d = zeros(1,numberOfParticles);
averageMsd = zeros( 1, longestTrack );
averageMsdCount = zeros( 1, longestTrack);
for ii = 1:numberOfParticles
if sum(particlesToSkip==ii)==0
numberOfMsds = length(particles{ii}.msd);
d(ii) = particles{ii}.diffusionCoefficient;
averageMsd(1:numberOfMsds) = averageMsd(1:numberOfMsds) + particles{ii}.msd;
averageMsdCount(1:numberOfMsds) = averageMsdCount(1:numberOfMsds) + 1;
end
end
figure( FigureHandle );
averageMsd = averageMsd ./ averageMsdCount;
loglog((1:longestTrack)/FrameRate, averageMsd, 'k', 'linewidth', 3);
ylabel('MSD (m^2)');
xlabel('Time (s)');
title('MSD versus Time Interval');
%hold off;
out = struct();
out.diffusionCoefficient = mean(d);
out.diffusionCoefficientUncertainty = std(d)/sqrt(length(d));
out.particles = particles;
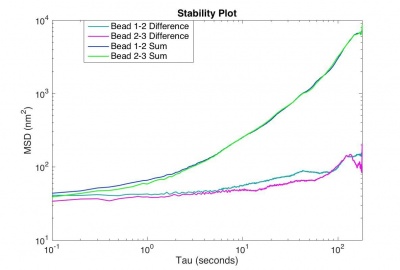
To characterize the performance of your microscope for particle tracking, you will make a 180 s movie of fixed, 0.84 μm microspheres with your microscope and plot the mean squared displacement of pairs of particles in the movie.
Stability of microscope for particle tracking
Once you have developed and tested your code, use the following procedure to measure the location precision of your microscope:
- Bring a slide with fixed beads into focus.
- Choose a field of view in which you can see at least 3 beads using the 40× objective.
- Limit the field of view to only those beads by choosing a region of interest (ROI) in UsefulImageAcquisition tool. (Otherwise, you will have a gigantic movie that is hard to work with.)
- Acquire a movie of beads for 3 minutes at a frame rate of at least 10 fps.
- Use the code you developed to track the particles and plot MSD versus time interval.
The MSD for the difference trajectory of two fixed particles (why the difference trajectory?) should start out less than 100 nm2 and still be less than 1000 nm2 for t = 180 s. If the MSD is larger than that, refine your apparatus and methodology until you achieve that goal.
| |
Turn in the code you developed and a plot of MSD for sum and difference tracks versus time interval. |
- Overview
- Part 1: Make a fake image
- Part 2: Measure resolution
- Part 3: Track microspheres over time
Back to 20.309 Main Page