20.109(S24):M2D4
Contents
Introduction
Protocols
Part 1: Participate in Communication Lab workshop
Our communication instructor, Dr. Chiara Ricci-Tam, will join us today for a discussion on oral presentations.
Part 2: Examine clone sequencing results
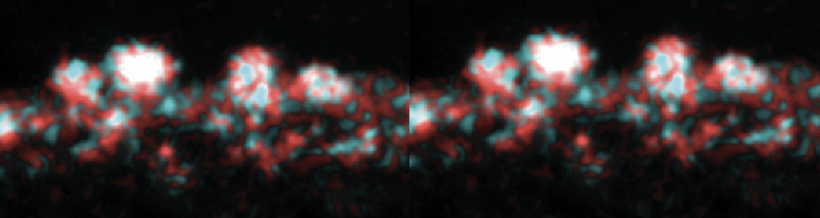
Your goal today is to analyze the sequencing data for you two YSD peptide clones - two independent colonies from your cloning reaction - and then decide which colony to proceed with to engineer yeast to become a cadmium sink.
Retrieve cloning sequence results from Genewiz
- Your sequencing data is available from Genewiz. For easier access, the information was uploaded to the [20.109(S24):Class_data Class Data tab].
- Download the zip folder with your team sequencing results and confirm that there are 8 files saved in the folder.
- For each sequencing reaction, you should have one .ab1 file and one .seq file.
- Open one of the .ab1 files.
- This file contains the chromatogram for your sequencing reaction. Scroll through the sequence and ensure that the peaks are clearly defined and evenly spaced. Low signal (or peaks) or stacked peaks can provide incorrect base assignments in the sequence.
- Open one of the .seq files.
- This file contains the base assignments for your sequencing reaction. The bases are assigned by the software from the chromatogram sequence.
- The start of the a sequencing reaction result often contains several Ns, which indicates that the software was unable to assign a basepair.
In your laboratory notebook, complete the following:
- Given the chromatogram result, why might the software assign Ns in the start of the sequence?
- Visually inspect the chromatograms for all of your sequencing results.
- Do the peaks appear clearly defined or is there overlap? What might this indicate about the quality of your sequencing results?
- Do the peaks extend above the background signal? What might this indicate about the quality of your sequencing results?
Confirm insertion sequence using SnapGene
You should align your sequencing data with a known sequence, in this case the DNA sequence encoding your peptide of interest from M2D1, to identify a successful insertion. There are several web-based programs for aligning sequences and still more programs that can be purchased. The steps for using SnapGene are below. Please feel free to use any program with which you are familiar.
- Generate a new DNA file that contains the insertion sequence you generated on M2D1.
- Generate an additional new DNA file that contains the results from the sequencing reaction completed by Genewiz.
- For each sequencing result you should generate a distinct new DNA file. Remember you should have a forward and reverse sequencing result for each of your clones!
- Paste the sequence text from your sequencing run into the new DNA file window. If there were ambiguous areas of your sequencing results, these will be listed as "N" rather than "A" "T" "G" or "C" and it's fine to include Ns in the query.
- The start and end of your sequencing may have several Ns. In this case it is best to omit these Ns by pasting only the 'good' sequence that is flanked by the ambiguous sequence.
- To confirm the mutation sequence in your clones, open one of the forward sequencing results files generated in the previous step.
- Select 'Tools' --> 'Align to Reference DNA Sequence...' --> 'Align Full Sequences...' from the toolbar.
- In the window, select the file that contains the insertion oligo sequence and click 'Open'.
- A new window will open with the alignment of the two sequences. The top line of sequence shows the results of the sequencing reaction and the bottom line shows the oligo you designed.
- Are there any discrepancies or differences between the two sequences? Scroll through the entire alignment to check the full sequencing result and note any basepair changes.
- Follow the above steps to examine all of your sequencing results. Remember: you used a forward and a reverse primer to check for insertion.
- From the alignments, determine which Fet_mutant clone has the correct point mutation.
- If both clones contain the correct sequence choose either yeast transformation to use in the rest of your experiments. If only one is correct, then this is the transformant you will use. If neither of your plasmids carry the appropriate mutation, talk to your Instructor.
In your laboratory notebook, complete the following:
- Attach a screenshot for each alignment.
- Record which clone contains the correct point mutation.
Part 3: Accuri Flow Cytometry of Stained Yeast Samples
To prepare for analysis, the Accuri flow cytometer needs to be flushed with fluid to remove any contamination from previous samples. Your samples will then be filtered one by one into round bottom analyzer tubes and placed under the cytometer sample injection port (SIP). The machine will then suck up the sample, and produce the dot plots. The Accuri will record the data from each laser and filter for every cell. The gating that the instructor performs will simply help visualize the samples as we perform the experiment but all the data will be saved automatically for post-processing analysis.
For more details on using the BD Accuri C6 flow cytometer, please watch the video tutorial linked here: Flow Cytometry. The steps are detailed below so you can follow along!
- Turn machine on. MilliQ water (ultra-pure water) should be in a tube on the sample injection port (SIP).
- Visually check waste containers and buffers.
- Open BD AccuriC6 software on the computer.
- To clean the machine, place a round bottom tube with cleaning solution on the SIP and under Run settings, choose "Run with Limits" and set 5 minutes and click RUN.
- Once the 5min cleaning is complete, place an empty tube on the SIP and under Run settings, click the backflush button to rinse the system.
- Place a fresh tube with 2ml of MilliQ water and run for 2min.
- You are now ready to analyze your samples.
- Place one filter cap tube in a rack and take your unstained yeast sample from the ice bucket.
- Invert the yeast microcentrifuge tube once, take the P1000 pipet and take up the yeast sample. Place the pipet tip flush against the filter cap and push the yeast sample through the 35um mesh into the round bottom tube.
- Remove the filter cap off of the yeast sample, take the MilliQ water tube off of the SIP and place the tube with filtered yeast on the SIP.
- Choose the analysis parameters in the software under Run settings. Run with limits: 10,000 events, instead of a time limit.
- Choose an analysis cell in the software with no events recorded and hit the RUN button.
- Unused cells are white (Example, C1) and cells with stored data are blue (Example, A1).
- We will detail gating and fluorescent analysis on M1D7.
- As the analysis proceeds, prepare your next yeast sample according to the steps above.
- Data is collected in individual cells in the software. Make sure to move the event record to a new cell between samples. Hit RUN each time you start a new sample analysis.
- Either name the cells with experimental details or write in your notes which samples are analyzed in each cell.
- Once all of the samples are analyzed, place a round bottom tube of Decon solution (10% bleach) on the SIP and run for 2min.
- The flow cytometer is now ready for the next user.
In your laboratory notebook, complete the following:
- XXX
Reagents list
Next day: Analyze flow cytometry and set up cadmium sequestration experiment