Difference between revisions of "20.109(S23):M2D1"
Becky Meyer (Talk | contribs) |
Becky Meyer (Talk | contribs) (→Protocols) |
||
| Line 14: | Line 14: | ||
==Protocols== | ==Protocols== | ||
| − | ===Part 1: Review | + | ===Part 1: Review Fet4 reference article=== |
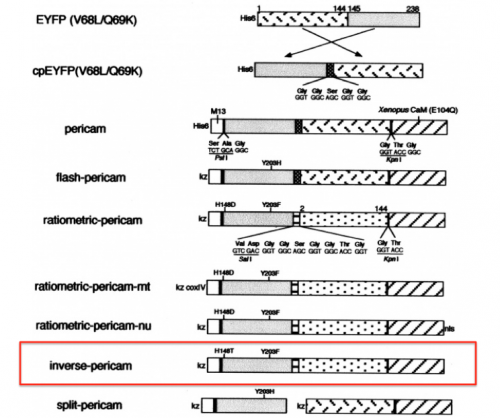
[[Image:Sp16 M1D2 pericam variants.png|thumb|right|500 px|'''Schematics of pericam variants.''' Representations of Ca<sup>2+</sup>-sensitive reporter constructs. In this module, your research will explore the properties of the inverse pericam construct (boxed in red). Image modified from Figure 1 of Nagai ''et. al.'', (2001) ''Proc. Natl. Acad. Sci.'' 98:3197.]] | [[Image:Sp16 M1D2 pericam variants.png|thumb|right|500 px|'''Schematics of pericam variants.''' Representations of Ca<sup>2+</sup>-sensitive reporter constructs. In this module, your research will explore the properties of the inverse pericam construct (boxed in red). Image modified from Figure 1 of Nagai ''et. al.'', (2001) ''Proc. Natl. Acad. Sci.'' 98:3197.]] | ||
| Line 29: | Line 29: | ||
*Why is it important / useful to construct a calcium sensor? Why is it useful / important to construct multiple calcium sensors with differing functionality / activity? | *Why is it important / useful to construct a calcium sensor? Why is it useful / important to construct multiple calcium sensors with differing functionality / activity? | ||
| − | |||
| − | + | ===Part 2: Examine Fet4 structural elements=== | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | ===Part | + | |
In the previous section you reviewed primary scientific literature to locate important features in the IPC sequence. Now you will examine 3D representations of CaM to visualize those features more closely. | In the previous section you reviewed primary scientific literature to locate important features in the IPC sequence. Now you will examine 3D representations of CaM to visualize those features more closely. | ||
| Line 109: | Line 84: | ||
**What additional regions might be interesting targets for mutagenesis? Why? | **What additional regions might be interesting targets for mutagenesis? Why? | ||
**What additional mutations do you think might impact the activity of IPC (be specific, what amino acid will replace what amino acid?). Do you hypothesize that this mutation will increase or decrease the affinity of calcium binding? Why? Do you hypothesize that this mutation will increase or decrease the cooperativity of calcium binding? Why? | **What additional mutations do you think might impact the activity of IPC (be specific, what amino acid will replace what amino acid?). Do you hypothesize that this mutation will increase or decrease the affinity of calcium binding? Why? Do you hypothesize that this mutation will increase or decrease the cooperativity of calcium binding? Why? | ||
| + | |||
| + | ===Part 4: Identify amino acid substitution target for new IPC design=== | ||
| + | Your first task for today is to review the data analysis you completed on M3D3 and decide which Variant IPC data you will consider when designing your Variant IPC. After you choose which amino acid you think is the best target for altering affinity / cooperativity, consider what amino acid you want to include instead. | ||
| + | |||
| + | In Part 5, you will generate the primers that can be used to incorporate a specific amino acid substitution to create your Variant IPC! | ||
| + | |||
| + | <font color = #4a9152 >'''In your laboratory notebook,'''</font color> complete the following: | ||
| + | *What amino acid will you target using SDM? At what position is this amino acid located in the protein sequence? What amino acid will be incorporated in its place? | ||
| + | *Provide the rational for your design choice. | ||
| + | **Why do you think the target amino acid you selected will alter affinity / cooperativity? | ||
| + | **How do you think the amino acid substitution will alter affinity / cooperativity? | ||
| + | |||
| + | ===Part 5: Design primers for site-directed mutagenesis=== | ||
| + | |||
| + | It is not experimentally efficient, or entirely plausible, to pick out and modify a single amino acid residue in inverse pericam post-translationally. Instead researchers genetically encode for amino acid substitutions by incorporating mutations in the DNA sequence. This is accomplished by making changes to the basepairs of a gene of interest that was cloned into a plasmid. Then the plasmid with the mutated gene is amplified using bacterial cells. | ||
| + | |||
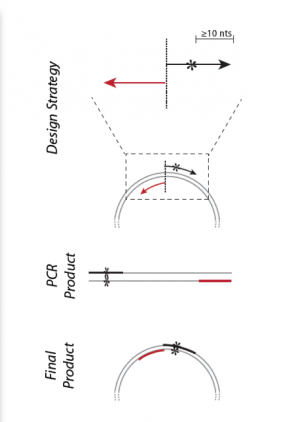
| + | [[Image:Sp16 M1D2 primer design schematic.png|thumb|right|300px| '''Schematic for mutating gene sequences in plasmids using SDM technique.''' Image modified from Q5 Site-Directed Mutagenesis Kit Manual published by NEB.]]To incorporate a mutation at a specific location in the DNA sequence, synthetic primers can be used in a technique referred to site-direction mutagenesis (see figure on the right). Primer design for site-directed mutagenesis, or SDM, is quite straightforward: the forward primer introduces a mutation into the coding strand. Both non-mutagenic and mutagenic amplification require cycles of DNA melting, annealing, and extension. | ||
| + | |||
| + | Primers used in SDM must meet several design criteria to ensure specificity and efficiency. Consider the following design guidelines for mutagenesis primers: | ||
| + | |||
| + | *Desired mutation (1-2 bp) must be present in the middle of the forward primer. | ||
| + | *Forward and reverse primers should 'face' away from the mutation and be 'back-to-back' when annealed to the template. | ||
| + | *Primers should be 25-45 bp long. | ||
| + | *G/C content of > 40% is desired. | ||
| + | *Both primers should terminate in at least one G or C base. | ||
| + | *Melting temperature should exceed 78°C, according to: | ||
| + | **T<sub>m</sub> = 81.5 + 0.41 (%GC) – 675/N - %mismatch | ||
| + | **where N is primer length and the two percentages should be integers | ||
| + | |||
| + | To demonstrate primer design, the illustration below uses S101L, which is an uninteresting mutation but a helpful example: | ||
| + | |||
| + | Residue 101 of calmodulin is serine, encoded by the AGC codon. This is residue 379 with respect to the entire inverse pericam construct, | ||
| + | and we can find it and some flanking code in the DNA sequence from Part 2: | ||
| + | |||
| + | <font face="courier"> | ||
| + | <small> | ||
| + | |||
| + | 361 (5') GAG GAA ATC CGA GAA GCA TTC CGT GTT TTT GAC AAG GAT GGG AAC GGC TAC ATC AGC GCT (3') | ||
| + | |||
| + | 381 (5') GCT CAG TTA CGT CAC GTC ATG ACA AAC CTC GGG GAG AAG TTA ACA GAT GAA GAA GTT GAT (3') | ||
| + | </small> | ||
| + | </font> | ||
| + | |||
| + | To change from serine to leucine, one might choose TTA, TTG, or CTN (wherer N = T, A, G, or C). Because CTC requires only two mutations (rather than three as for the other options), we choose this codon. | ||
| + | |||
| + | Now we must keep >10 bp of sequence on each side in a way that meets all our requirements. To quickly find G/C content and see secondary structures, look at the [https://www.idtdna.com/pages/tools/oligoanalyzer IDT website]. (Note that the T<sub>m</sub> listed at this site is '''''not''''' one that is relevant for mutagenesis.) | ||
| + | |||
| + | Ultimately, your forward primer might look like the following, which has a T<sub>m</sub> of almost 81°C, and a G/C content of ~58%. | ||
| + | |||
| + | <font face="courier"> | ||
| + | 5’ GG AAC GGC TAC ATC CTC GCT GCT CAG TTA CGT CAC G 3' | ||
| + | </font><br> | ||
| + | |||
| + | The reverse primer is the inverse complement of a sequence just preceding the forward primer in the IPC gene. The forward and reverse primers are set up back-to-back. | ||
| + | |||
| + | Luckily, online tools are available to assist with SDM primer design. Today you will use NEBaseChanger (provided by NEB) to design your mutagenic primers. | ||
| + | #Go to the [http://nebasechanger.neb.com/ NEBaseChanger] site and click 'Please enter a new sequence to begin.' | ||
| + | #*A new window will open. | ||
| + | #Copy and paste the WT IPC sequence. | ||
| + | #*This sequence should be saved in SnapGene from the M3D1 exercise. Alternatively, you can copy the sequence from the word document attached to the M3D1 wiki page. | ||
| + | #Confirm that the 'Substitution' option is selected. | ||
| + | #Highlight the basepairs you want to mutate using by scrolling through the sequence, or you can search the sequence by typing the basepairs into the 'Find' box. | ||
| + | #Type the new DNA sequence (the basepair(s) you want your forward mutagenic primer to incorporate into the IPC sequence) in the 'Desired Sequence' box. | ||
| + | #*Under the Result header, a diagram showing where your primers will anneal is provided. | ||
| + | #*Under the Required Primers header, the sequences for your forward primer and reverse primer are shown with the characteristics for each. | ||
| + | #<font color = #4a9152 >'''In your laboratory notebook,'''</font color> complete the following: | ||
| + | #*Include a screen capture of the information provided in the Result and Required Primers sections. | ||
| + | #*Use the guidelines provided above to examine the mutagenesis primers designed by NEBaseChanger. Do the primers meet the design criteria? | ||
==Navigation links== | ==Navigation links== | ||
Next day: [[20.109(S23):M2D2 |Perform site-directed mutagenesis]] <br> | Next day: [[20.109(S23):M2D2 |Perform site-directed mutagenesis]] <br> | ||
Previous day: [[20.109(S21):M1D8 |Organize Data summary figures and results]] <br> | Previous day: [[20.109(S21):M1D8 |Organize Data summary figures and results]] <br> | ||
Revision as of 19:01, 1 February 2023
Contents
Introduction
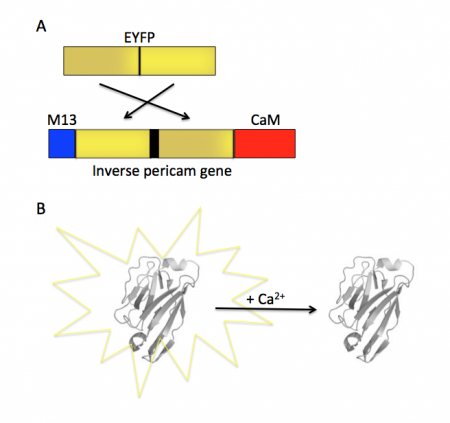
Today you will familiarize yourself with the recombinant protein IPC and its constituent parts. The fluorescent component of IPC is an enhanced yellow fluorescent protein (abbreviated EYFP), one of the many derivatives of green fluorescent protein (GFP). GFP is naturally produced by jellyfish and was cloned into other organisms in the early 1990’s. It has since been exploited as a genetically encodable reporter and mutagenized to vary its excitation and emission spectra. The other key component of inverse pericam is the protein calmodulin (CaM), a natural calcium sensor that is present in all eukaryotes. Calmodulin has many ligands that it binds only in the presence of calcium ion, including the peptide fragment M13. This conditional specificity for M13 binding is enabled by the change in confirmation of CaM when bound to calcium.
Within inverse pericam, M13 and CaM are located at opposite ends, surrounding a permuted (i.e., rearranged) version of EYFP. In the absence of calcium, this EYFP exhibits strong fluorescence. However, when enough calcium is added to a solution of inverse pericam, CaM and M13 interact, disrupting the conformation and, as a result, the fluorescence of EYFP. The transition from bright to dim fluorescence occurs over a particular concentration range of calcium. The calcium concentration at which binding to CaM occurs (and fluorescence decreases) is referred to as the Kd and determined by the affinity of CaM to calcium. In addition, the interaction between CaM and calcium is impacted by cooperativity. CaM has four calcium binding sites. In cooperativity, the affinity of CaM for calcium is altered by how many calcium ions are already bound to the protein. The mutations you will examine today were designed in an effort to modify the calcium sensor portion of IPC in a manner that is likely to change the affinity and / or cooperativity for calcium ions.
To examine the modification that were made to IPC, we will use several protein analysis tools. Proteins are modular materials that may be described and examined at multiple levels of a structural hierarchy (from primary to quaternary in the classical paradigm). Primary structure refers to a protein’s amino acid sequence, which might reveal a cluster of charged residues or a pattern of alternating polar and nonpolar residues. One cannot predict off-hand the conformation of a protein merely from its linear sequence; however, due to rotational flexibility of bonds and non-covalent interactions between non-adjacent amino acids (as well as covalent disulfide bonds) some structural characteristics can be inferred. Because many proteins have structural motifs in common (e.g., alpha helices and beta sheets at the secondary level, or leucine-rich repeats at the tertiary level), which ultimately arise from the amino acid sequences, databases can be useful for making predictions about proteins with known amino acid sequences but unknown structures.
Protocols
Part 1: Review Fet4 reference article
Previous 109ers generated the data you will analyze in this module by changing specific amino acids in the IPC protein sequence. The goal of this directed mutagenesis approach was to alter the interaction between IPC and calcium such that the affinity / cooperativity was improved. Before we examine the effect of these mutations on the activity of IPC, we will first review how the original IPC sensor was constructed.
With your partner, review the information regarding the development of calcium sensor variants in the paper by Nagai et. al. (attached here). Specifically, you will focus on the development and activity of flash pericam, radiometric pericam, and inverse pericam. The domain structures for these variants are shown in the schematic to the right.
In your laboratory notebook, complete the following:
- What is cpGFP? How was this molecule constructed? Why might this molecule be a more useful tool than GFP?
- What is cpYFP? How does it differ from cpGFP?
- What is calmodulin? What is M13?
- Review the references cited by the authors or use other resources to provide a brief description of each.
- What are the critical mutations that were identified in flash pericam, radiometric pericam, and inverse pericam? How do the authors propose that these critical mutations are involved in the activity of the calcium sensor variants?
- Why is it important / useful to construct a calcium sensor? Why is it useful / important to construct multiple calcium sensors with differing functionality / activity?
Part 2: Examine Fet4 structural elements
In the previous section you reviewed primary scientific literature to locate important features in the IPC sequence. Now you will examine 3D representations of CaM to visualize those features more closely.
- You will examine the structure of CaM using the Protein Data Bank (PDB) (linked here). In this online database, the structures are organized according to PDB identification codes.
- For this exercise, you will look at the calcium-bound form of CaM.
- Enter "1CLL" into the search box at the top right corner of the PDB homepage.
- The landing page for the CaM structure includes background information on the source and reference for this protein structure.
- In your laboratory notebook, complete the following:
- What method was used to solve this protein structure? Perform a quick search to learn more about this method and provide a brief description.
- At what resolution was the structure solved? Perform a quick search to learn more about this concept and provide a brief description.
- What is the total weight of the structure?
- How many chains are included in this structure?
- Read the abstract for the reference article wherein this structure was first published. What are the features of the calcium-binding domains (lobes) as described by the authors?
- Under the structure shown on the left side of the window, click the 'Structure' link. A page showing the 'cartoon' structure of CaM will load. Using the tools to the right of this page you will be able to more closely examine the structure.
- First, let's orient ourselves on how to move / manipulate the protein structure.
- Place your cursor over the structure and while pressing down on your mouse / track pad, move the image to view the protein structure from different angles.
- To zoom-in on an area of the protein structure, place your cursor on the area of interest and double-click. When zoomed in single-click on a residue to get a more detailed view of the amino acids that are present in that area. The dotted lines represent bonds or salt bridges that exist between the elements in the amino acids.
- To zoom-out, single-click on the white space in the viewer window.
- To zoom-in or -out more gradually, use two fingers and drag in the up or down direction.
- To identify which amino acid residues are present in each position of the protein, hover your cursor over the protein. A box will appear in the lower right of the viewer window (see example to the right). Though most of the details here can be ignored, the information provided tells you that the highlighted residue is a valine (Val) at position 35 in the amino acid sequence.
- In your laboratory notebook, complete the following:
- What secondary structures are present in CaM?
- Compare the description of the features within the lobes provided by the authors to the protein structure. Screen capture a zoomed in view of one of the lobes and label the features.
- Next, let's consider the tools provided in the panel on the right of the page.
- The contents of the 'Components' tab are listed: Polymer, Ligand, Water, and Ion.
- Polymer refers the larger structures present, such as protein chains, DNA, or RNA.
- Ligand refers to any non-polymer structure, such as ligand binders, ATP, or co-factors that are not single atoms.
- Water refers to water.
- Ion refers to any lone elements that are associated with the structure.
- Use the 'eyeball' icon to the right of the component labels to remove / add the components to the image.
- In your laboratory notebook, complete the following:
- Does the CaM structure contain the Components listed? Answer yes or no for each Component type.
- What type of Component is calcium? Include screen shots of a binding loop with and without calcium present.
- Click on the 'Density' tab. Though we will not focus much on the details here, the electron density map is the actual data from the x-ray crystallography experiment used to solve the structure.
- Select '2Fo-Fc σ' from the options.
- Click the box to the right of 'Wireframe' such that this feature is activated (toggle to '✓ On').
- Click on a residue within the protein structure. This will zoom-in on that area and also layer a grid, or cage, over the area. The cage represents the electron density data that were captured via x-ray crystallography. The structural features and atoms within the CaM protein were modeled to match the density map, thus providing a best estimate of the protein structure. The resolution is related to how tight this cage is to the solved structure. Though a gross oversimplification, the relationship can be described as such: the fit of the cage to the solved structure is related to the angstrom value achieved via crystallography, the smaller the angstrom the better the resolution and thus the tighter the cage to the solved structure.
- Lastly, let's look at how calcium interacts with CaM!
- Move the protein structure such that you are able to achieve a clear view of the calcium ion in the first binding loop and double-click on one of the residues in the loop to zoom-in.
- Hint: hover over the amino acid residues to identify the N-terminus based on the residue numbers provided in the box at the lower right of the viewer window.
- Single-click on the calcium ion to visualize how it associates with the residues in the loop.
- It may be easier to view the bonds by removing the density information from the structure. To do this, click the 'eyeball' icon to the right of each of the options listed in the 'Density' tab. Alternatively, you can exit the viewer window and re-enter to return the default setting to '☓ Off'.
- In your laboratory notebook, complete the following:
- List the amino acids (from N- to C-terminus) that are in the binding loop.
- List the amino acids (from N- to C-terminus) that are shown to interact with calcium in the binding loop. How many bonds are formed with each of the listed amino acids?
- Provide the above information for each of the binding loops present in the CaM structure.
- It may also be interesting to consider how the amino acids that are not directly bound to calcium interact as this is important to maintaining the structural integrity of the binding loop.
- Identify the isoleucine (Ile) residue at position 27, then single-click to show the relevant binding information.
- To identify which amino acid residues are bound to Ile 27, hover your cursor over the dotted lines. A box will appear in the lower right of the viewer window (see example to the right). As before, most of the details here can be ignored, the information provided tells you that the highlighted bond is a hydrogen bond between the oxygen (O) of Ile 63 and the nitrogen (N) of Ile 27.
In your laboratory notebook, complete the following:
- Based on what you learned from the protein structure, revisit the questions answered when examining the sequence for CaM:
- What additional regions might be interesting targets for mutagenesis? Why?
- What additional mutations do you think might impact the activity of IPC (be specific, what amino acid will replace what amino acid?). Do you hypothesize that this mutation will increase or decrease the affinity of calcium binding? Why? Do you hypothesize that this mutation will increase or decrease the cooperativity of calcium binding? Why?
Part 4: Identify amino acid substitution target for new IPC design
Your first task for today is to review the data analysis you completed on M3D3 and decide which Variant IPC data you will consider when designing your Variant IPC. After you choose which amino acid you think is the best target for altering affinity / cooperativity, consider what amino acid you want to include instead.
In Part 5, you will generate the primers that can be used to incorporate a specific amino acid substitution to create your Variant IPC!
In your laboratory notebook, complete the following:
- What amino acid will you target using SDM? At what position is this amino acid located in the protein sequence? What amino acid will be incorporated in its place?
- Provide the rational for your design choice.
- Why do you think the target amino acid you selected will alter affinity / cooperativity?
- How do you think the amino acid substitution will alter affinity / cooperativity?
Part 5: Design primers for site-directed mutagenesis
It is not experimentally efficient, or entirely plausible, to pick out and modify a single amino acid residue in inverse pericam post-translationally. Instead researchers genetically encode for amino acid substitutions by incorporating mutations in the DNA sequence. This is accomplished by making changes to the basepairs of a gene of interest that was cloned into a plasmid. Then the plasmid with the mutated gene is amplified using bacterial cells.
Primers used in SDM must meet several design criteria to ensure specificity and efficiency. Consider the following design guidelines for mutagenesis primers:
- Desired mutation (1-2 bp) must be present in the middle of the forward primer.
- Forward and reverse primers should 'face' away from the mutation and be 'back-to-back' when annealed to the template.
- Primers should be 25-45 bp long.
- G/C content of > 40% is desired.
- Both primers should terminate in at least one G or C base.
- Melting temperature should exceed 78°C, according to:
- Tm = 81.5 + 0.41 (%GC) – 675/N - %mismatch
- where N is primer length and the two percentages should be integers
To demonstrate primer design, the illustration below uses S101L, which is an uninteresting mutation but a helpful example:
Residue 101 of calmodulin is serine, encoded by the AGC codon. This is residue 379 with respect to the entire inverse pericam construct, and we can find it and some flanking code in the DNA sequence from Part 2:
361 (5') GAG GAA ATC CGA GAA GCA TTC CGT GTT TTT GAC AAG GAT GGG AAC GGC TAC ATC AGC GCT (3')
381 (5') GCT CAG TTA CGT CAC GTC ATG ACA AAC CTC GGG GAG AAG TTA ACA GAT GAA GAA GTT GAT (3')
To change from serine to leucine, one might choose TTA, TTG, or CTN (wherer N = T, A, G, or C). Because CTC requires only two mutations (rather than three as for the other options), we choose this codon.
Now we must keep >10 bp of sequence on each side in a way that meets all our requirements. To quickly find G/C content and see secondary structures, look at the IDT website. (Note that the Tm listed at this site is not one that is relevant for mutagenesis.)
Ultimately, your forward primer might look like the following, which has a Tm of almost 81°C, and a G/C content of ~58%.
5’ GG AAC GGC TAC ATC CTC GCT GCT CAG TTA CGT CAC G 3'
The reverse primer is the inverse complement of a sequence just preceding the forward primer in the IPC gene. The forward and reverse primers are set up back-to-back.
Luckily, online tools are available to assist with SDM primer design. Today you will use NEBaseChanger (provided by NEB) to design your mutagenic primers.
- Go to the NEBaseChanger site and click 'Please enter a new sequence to begin.'
- A new window will open.
- Copy and paste the WT IPC sequence.
- This sequence should be saved in SnapGene from the M3D1 exercise. Alternatively, you can copy the sequence from the word document attached to the M3D1 wiki page.
- Confirm that the 'Substitution' option is selected.
- Highlight the basepairs you want to mutate using by scrolling through the sequence, or you can search the sequence by typing the basepairs into the 'Find' box.
- Type the new DNA sequence (the basepair(s) you want your forward mutagenic primer to incorporate into the IPC sequence) in the 'Desired Sequence' box.
- Under the Result header, a diagram showing where your primers will anneal is provided.
- Under the Required Primers header, the sequences for your forward primer and reverse primer are shown with the characteristics for each.
- In your laboratory notebook, complete the following:
- Include a screen capture of the information provided in the Result and Required Primers sections.
- Use the guidelines provided above to examine the mutagenesis primers designed by NEBaseChanger. Do the primers meet the design criteria?
Next day: Perform site-directed mutagenesis