20.109(S22):M2D5
Contents
Introduction
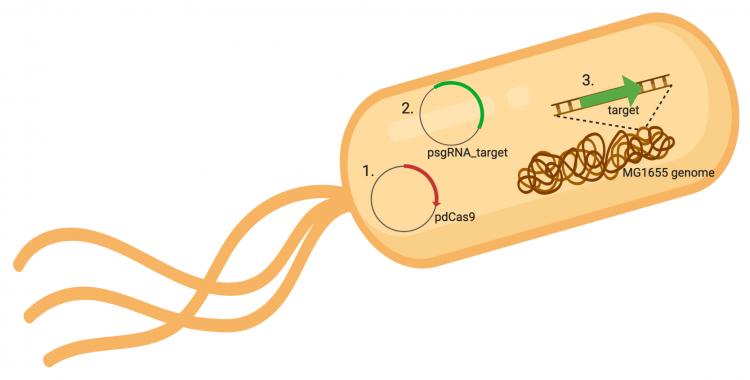
The CRISPRi system involves three genetic components: the pdCas9 plasmid (1 in image below), the psgRNA_target plasmid (2 in image below), and the targeted gene within the host genome (3 in image below). Though the targeted gene is native to the host genome, the plasmids must be transformed into the cell and maintained using antibiotic selection. Thus far in this module, we have discussed the CRISPRi plasmids as individual units, but now we will consider the system as a whole in the context of engineering gene expression.
In the previous laboratory session, you reviewed the procedure used to generate the psgRNA_target plasmids. Today we will discuss how the CRISPRi system (pdCasd9 and psgRNA_target) is transformed into E. coli and, once transformed into the bacterial cells, how the psgRNA_target and dCas9 are transcribed from the expression plasmids. Briefly, the promoter (pJ23119) driving expression of the sgRNA sequence in the gRNA_target plasmid is constitutively active. This means that transcription of the gRNA sequence specific to the target in the host genome is constitutive. Therefore, your sgRNA_target is always present in the MG1655 cells.
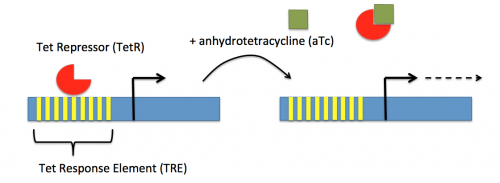
In contrast, expression of the gene encoding dCas9 within pdCas9 is regulated by an inducible promoter (pLtetO-1). An inducible promoter is 'off' unless the appropriate molecule is present to relieve repression. In the case of the CRISPRi system, expression of the gene encoding dCas9 is inhibited due to the use of a tet-based promoter construct. Tet is shorthand for tetracycline, which is an antibiotic that inhibits protein synthesis through preventing the association between charged aminoacyl-tRNA molecules and the A site of ribosomes. Bacterial cells that carry the tet resistance cassette are able to survive exposure to tetracycline by expressing genes that encode an efflux pump that 'flushes' the antibiotic from the bacterial cell. To conserve energy, the tet system is only expressed in the presence of tetracyline. In the absence of tetracycline, a transcription repressor protein (TetR) is bound to the promoter upstream of the tet resistance cassette genes. When tetracycline is present, the molecule binds to TetR causing a confirmational change that results in TetR 'falling off' of the promoter. In the CRISPRi system, the tet-based promoter construct upstream of the gene that encodes dCas9 is 'off' unless anhydrotetracyline (aTc), an analog of tetracyline, is added to the culture media. Why is it important to use an analog rather than the actual antibiotic?Taken together, the sgRNA_target is constitutively transcribed and, as stated above, always present. The dCas9 protein is only present when aTc is added. Thus, gene expression is only altered when aTc is present. In this, when dCas9 is expressed it forms a complex with the sgRNA_target. The sgRNA target then 'seeks out' the target within the host genome. When the targeted sequence is recognized, the complex binds and acts as a 'roadblock' by prohibiting RNAP access to the sequence. Because the targeted gene is not able to be transcribed, the protein encoded by that gene is not synthesized. In our experiments, we hypothesize that the absence of specific proteins, or enzymes, involved in anaerobic fermentative metabolism will increase the yield of ethanol.
Protocols
Part 3: Research CRISPRi system
Review and discuss the Introduction and the first Results sub-section ('A minimal CRISPRi system consists of a single protein and RNA and can effectively silence transcription initiation and elongation') from the following journal article with your laboratory partner:
Lei et al. "Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression." Cell. (2013) 152:1173-1183.
In this article, the researchers describe a modified CRISPR system, referred to as CRISPR interference (CRISPRi), that inhibits transcription of genes that are targeted using single guide RNA (sgRNA) molecules. The goal of this exercise is to understand how the sgRNA molecules are used to target specific genes. In the CRISPRi system, sgRNA forms a complex with dCas9 and this complex binds to the sequence in the genome that is complementary to the sgRNA molecule. Thus, the sgRNA targets a specific gene sequence in the host genome. When the sgRNA / dCas9 complex binds, transcription of the gene is inhibited.
Transcription can be blocked by targeting sgRNA molecules to the promoter or coding region of a specific gene. The promoter is the region upstream of the start codon and contains the RNAP binding site, or the -10 and -35. When the sgRNA / dCas9 complex binds to the promoter, transcription initiation is inhibited because RNAP is unable to bind to the promoter. The coding region refers to the gene sequence. When the sgRNA is targeted to the coding region, transcription elongation is stalled because RNAP is unable to traverse beyond the location of the sgRNA / dCas9 complex.
In your laboratory notebook, complete the following with your partner:
- Briefly, describe how the CRISPRi system is different than the native CRISPR system that was discussed during lecture.
- Hint: Which two components were modified? How?
- In the first paragraph of the Results sub-section, what are the researchers testing?
- Describe the red fluorescent protein (mRFP)-based reporter system.
- Where was the mRFP gene sequence inserted?
- What does this system enable the researchers to test?
- For Fig. 2C, describe the schematic of the mRFP-based reporter system (top of the panel).
- What are each of the components and what is the purpose of each?
- What does it mean if RFP is expressed from this construct? If RFP is not expressed from this construct?
- For Fig. 2C, explain the data that are shown (bottom of the panel).
- What is the control?
- What do these data tell you about how to best target the coding region of a gene using sgRNA molecules and the CRISPRi system?
- For Fig. 2D, describe the schematic of the experimental approach (top of panel).
- How does this experiment relate to what is shown in Fig. 2C?
- The output from this experimental approach is RFP. Where is the RFP insert? How does this approach relate to the mRFP-based reporter system?
- For Fig. 2D, explain the data that are shown (bottom of the panel).
- What is the control?
- What do these data tell you about how to best target the promoter region of a gene using sgRNA molecules and the CRISPRi system?
- Lastly, given the results from this article what is the best approach for targeting the promising candidate you identified above in Part 1?
- Should you design an sgRNA that targets, or binds, the promoter or the coding region? The template or the non-template strand?
- Where in the promoter or coding region should the sgRNA bind (ie the -35 or the middle of the gene)?
Prepare media for ethanol yield experiment
To test the effect of sgRNA_target on increasing the production of ethanol, the co-transformed E. coli MG1655 cells both with and without oxygen. Remember from lecture that cells grown anaerobically should produce more fermentation products; however, the goal of using the CRISPRi system is to further enhance the production of ethanol by manipulating gene expression of an enzyme in the anaerobic fermentative pathway.
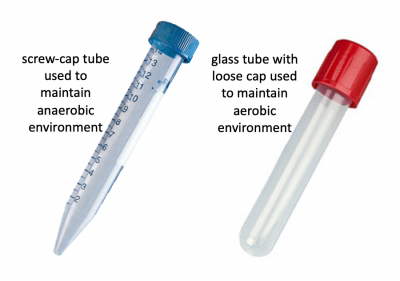
Before we look at the specific conditions that were tested, let's first review how anaerobic and aerobic cultures were maintained. Rather than using anaerobic chambers or gas replacement, a much simpler technique was employed to omit oxygen from the cultures for this experiment. Because screw-cap tubes maintain a tight seal, cultures can be maintained in a low-O2 environment. For this technique cultures are grown at least 24 hrs to ensure the following: 1. that the oxygen present at the time of inoculation is depleted by the growing cells and 2. that the cells grow in the O2-depleted environment long enough to undergo anaerobic fermentative metabolism. Aerobic cultures were maintained using standard glass test tubes with loose-fitting caps. Both culture tubes are shown in the image to the right.In total, eight conditions were tested for each sgRNA_target:
- MG1655 +O2 -aTc (glass tube)
- MG1655 -O2 -aTc (screw-cap tube)
- MG1655 +O2 +aTc (glass tube)
- MG1655 -O2 +aTc (screw-cap tube)
- MG1655 +CRISPRi +O2 -aTc (glass tube)
- MG1655 +CRISPRi -O2 -aTc (screw-cap tube)
- MG1655 +CRISPRi +O2 +aTc (glass tube)
- MG1655 +CRISPRi -O2 +aTc (screw-cap tube)
All cultures were prepared in 5 mL LB broth containing 25 μg/mL of chloramphenicol and 100 μg/mL of ampicillin.
In your laboratory notebook, complete the following:
- For each of the conditions that were tested, answer the following questions:
- Is the cell culture using anaerobic fermentative metabolism?
- Do the cells contain the CRISPRi system?
- Is the CRISPRi system active (are the components expressed)?
- Which culture conditions provide controls? What does each control indicate / validate?
- In which culture condition do you expect to see the lowest yield of ethanol? Why?
- In which culture condition do you expect to see the highest yield of ethanol? Why?
Reagents list
Next day: Complete CRISPRi experiment and measure fermentation products