Difference between revisions of "20.109(S22):M2D5"
Noreen Lyell (Talk | contribs) (Created page with "<div style="padding: 10px; width: 820px; border: 5px solid #434a43;"> {{Template:20.109(S22)}} ==Introduction== The CRISPRi system involves three genetic components: the p...") |
(No difference)
|
Revision as of 14:05, 21 January 2022
Introduction
The CRISPRi system involves three genetic components: the pdCas9 plasmid (1 in image below), the psgRNA_target plasmid (2 in image below), and the targeted gene within the host genome (3 in image below). Though the targeted gene is native to the host genome, the plasmids must be transformed into the cell and maintained using antibiotic selection. Thus far in this module, we have discussed the CRISPRi plasmids as individual units, but now we will consider the system as a whole in the context of engineering gene expression.
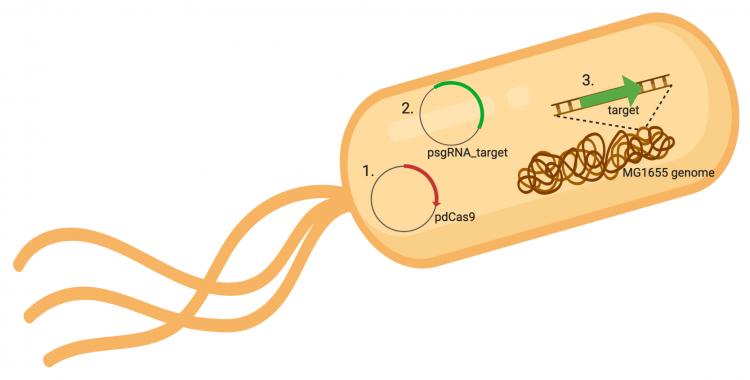
In the previous laboratory session, you reviewed the procedure used to generate the psgRNA_target plasmids. Today we will discuss how the CRISPRi system (pdCasd9 and psgRNA_target) is transformed into E. coli and, once transformed into the bacterial cells, how the psgRNA_target and dCas9 are transcribed from the expression plasmids. Briefly, the promoter (pJ23119) driving expression of the sgRNA sequence in the gRNA_target plasmid is constitutively active. This means that transcription of the gRNA sequence specific to the target in the host genome is constitutive. Therefore, your sgRNA_target is always present in the MG1655 cells.
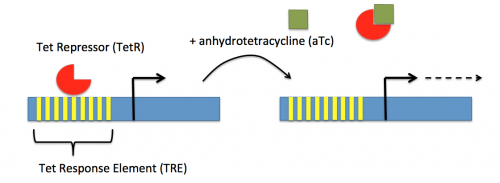
In contrast, expression of the gene encoding dCas9 within pdCas9 is regulated by an inducible promoter (pLtetO-1). An inducible promoter is 'off' unless the appropriate molecule is present to relieve repression. In the case of the CRISPRi system, expression of the gene encoding dCas9 is inhibited due to the use of a tet-based promoter construct. Tet is shorthand for tetracycline, which is an antibiotic that inhibits protein synthesis through preventing the association between charged aminoacyl-tRNA molecules and the A site of ribosomes. Bacterial cells that carry the tet resistance cassette are able to survive exposure to tetracycline by expressing genes that encode an efflux pump that 'flushes' the antibiotic from the bacterial cell. To conserve energy, the tet system is only expressed in the presence of tetracyline. In the absence of tetracycline, a transcription repressor protein (TetR) is bound to the promoter upstream of the tet resistance cassette genes. When tetracycline is present, the molecule binds to TetR causing a confirmational change that results in TetR 'falling off' of the promoter. In the CRISPRi system, the tet-based promoter construct upstream of the gene that encodes dCas9 is 'off' unless anhydrotetracyline (aTc), an analog of tetracyline, is added to the culture media. Why is it important to use an analog rather than the actual antibiotic?Taken together, the sgRNA_target is constitutively transcribed and, as stated above, always present. The dCas9 protein is only present when aTc is added. Thus, gene expression is only altered when aTc is present. In this, when dCas9 is expressed it forms a complex with the sgRNA_target. The sgRNA target then 'seeks out' the target within the host genome. When the targeted sequence is recognized, the complex binds and acts as a 'roadblock' by prohibiting RNAP access to the sequence. Because the targeted gene is not able to be transcribed, the protein encoded by that gene is not synthesized. In our experiments, we hypothesize that the absence of specific proteins, or enzymes, involved in anaerobic fermentative metabolism will increase the yield of ethanol.