Difference between revisions of "20.109(S22):M1D4"
Noreen Lyell (Talk | contribs) (→Part 3: Visualize TDP43-RRM12 protein using polyacrylamide gel electrophoresis (PAGE)) |
(→Part 3: Measure protein concentration) |
||
| (17 intermediate revisions by 2 users not shown) | |||
| Line 23: | Line 23: | ||
Our communication instructor, Dr. Prerna Bhargava, will join us today for a discussion on crafting data figures and legends. | Our communication instructor, Dr. Prerna Bhargava, will join us today for a discussion on crafting data figures and legends. | ||
| − | ===Part 2 | + | ===Part 2: Visualize TDP43-RRM12 protein using polyacrylamide gel electrophoresis (PAGE)=== |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
As discussed above, SDS-PAGE is used to evaluate the purity protein. It is also possible to track the purification process using SDS-PAGE. For this, several aliquots were collected during the protein purification and concentration procedures. Each of these aliquots will be electrophoresed and the presence / absence of the protein of interest in each of the aliquots will provide information on the success of the purification. | As discussed above, SDS-PAGE is used to evaluate the purity protein. It is also possible to track the purification process using SDS-PAGE. For this, several aliquots were collected during the protein purification and concentration procedures. Each of these aliquots will be electrophoresed and the presence / absence of the protein of interest in each of the aliquots will provide information on the success of the purification. | ||
| − | One important note to consider is that the expected size of the TDP43-RRM12 product is dependent on the features that were added during the cloning process (review your notes from M2D1 prelab discussion!). The molecular weight of TDP43-RRM12 is ~20 kDa. Each of the features adds 'weight' to the product: SnapTag is ~20 kDa, 6x-His | + | One important note to consider is that the expected size of the TDP43-RRM12 product is dependent on the features that were added during the cloning process (review your notes from M2D1 prelab discussion!). The molecular weight of TDP43-RRM12 is ~20 kDa. Each of the features adds 'weight' to the product: SnapTag is ~20 kDa, 3C recognition site is ~1 kDa, MBP is ~40 kDa, and 6x-His is ~1 kDa. As you evaluate each lane of the SDS-PAGE results below, critically think about what the expected band size is for each of the samples. Review the M1D3 prelab slides for the orientation of the components that makeup the purified product. |
| − | + | ||
| − | + | ||
<font color = #0d368e>'''To see a closer view of loading protein sample into an SDS-PAGE gel, please watch the video linked here: </font color>[[https://www.dropbox.com/s/sno0xghpte8moby/Loading%20a%20protein%20gel.mp4?dl=0 Loading samples into a protein gel]]. | <font color = #0d368e>'''To see a closer view of loading protein sample into an SDS-PAGE gel, please watch the video linked here: </font color>[[https://www.dropbox.com/s/sno0xghpte8moby/Loading%20a%20protein%20gel.mp4?dl=0 Loading samples into a protein gel]]. | ||
#Retrieve the aliquots that were collected during the protein purification and concentration procedures. | #Retrieve the aliquots that were collected during the protein purification and concentration procedures. | ||
| − | #*In addition to the aliquots that were | + | #*In addition to the aliquots that were prepared in the previous laboratory session, you will also include the desalted protein in the SDS-PAGE experiment. To do this, transfer 30 μL from the 15 mL conical with your desalted protein into a microcentrifuge tube. |
#To 30 μL of each aliquot, add 6 μL of Laemmli sample buffer. | #To 30 μL of each aliquot, add 6 μL of Laemmli sample buffer. | ||
#*Prepare the cell pellet by adding 30 μL of water to a fresh microcentrifuge tube, then dip a clean pipette tip in the bacterial pellet and swirl into the water. Lastly, add 6 μL of Laemmli sample buffer. | #*Prepare the cell pellet by adding 30 μL of water to a fresh microcentrifuge tube, then dip a clean pipette tip in the bacterial pellet and swirl into the water. Lastly, add 6 μL of Laemmli sample buffer. | ||
| − | #Boil all samples for 5 min in the water bath located | + | #Boil all samples for 5 min in the water bath (set at 100 °C) located at the front laboratory bench. |
| − | #*Secure the tubes with the cap-locks located | + | #*Secure the tubes with the cap-locks located next to baths to ensure that the caps do not pop open during the boiling step as this will result in your sample evaporating from the tube. |
| − | # | + | #Vortex and centrifuge samples for 15 seconds each to resuspend and collect samples in the bottoms of the tubes, respectively. Vortex and table-top mini centrifuge are at front bench. |
#You will load all samples and a molecular weight standard. | #You will load all samples and a molecular weight standard. | ||
#Load the total volume for the samples and 10 μL of the ladder into the polyacrylamide gel. | #Load the total volume for the samples and 10 μL of the ladder into the polyacrylamide gel. | ||
| Line 68: | Line 47: | ||
#*Be careful that the gel does not fall into the sink! | #*Be careful that the gel does not fall into the sink! | ||
#Repeat Steps #10-11 a total of 3 times. | #Repeat Steps #10-11 a total of 3 times. | ||
| − | # | + | #Cover the gel with the BioSafe Coomassie in the staining box and incubate on the rotating table at room temperature overnight. |
#Empty the BioSafe Coomassie into the appropriate waste container in the chemical fume hood. | #Empty the BioSafe Coomassie into the appropriate waste container in the chemical fume hood. | ||
#*Be careful that the gel does not fall into the waste container! | #*Be careful that the gel does not fall into the waste container! | ||
| Line 75: | Line 54: | ||
#*Replace the dH<sub>2</sub>O every 30 min. | #*Replace the dH<sub>2</sub>O every 30 min. | ||
#Image gel using the white light setting, not the UV light setting, of the gel documentation station. | #Image gel using the white light setting, not the UV light setting, of the gel documentation station. | ||
| − | |||
<font color = #4a9152 >'''In your laboratory notebook,'''</font color> complete the following: | <font color = #4a9152 >'''In your laboratory notebook,'''</font color> complete the following: | ||
*Consider the expected results for each lane in the polyacrylamide gel. | *Consider the expected results for each lane in the polyacrylamide gel. | ||
| − | **Do you ''expect'' to see the TDP43-RRM12 protein in the lane? If yes, what the expected size? If no, why not? | + | **Do you ''expect'' to see the TDP43-RRM12 protein in the lane? If yes, what is the expected size? If no, why not? |
**Do you ''expect'' to see other cellular proteins in the lane? If yes, why? If no, why not? | **Do you ''expect'' to see other cellular proteins in the lane? If yes, why? If no, why not? | ||
| − | ===Part | + | ===Part 3: Measure protein concentration=== |
| + | |||
SDS-PAGE enables you to visualize the presence of protein and provides information concerning the purity of your protein sample(s). Though comparing SDS-PAGE band intensities between samples and molecular weight standards gives an estimate of protein concentration, using a standard curve generated from samples of known protein concentration is a more precise method for measuring protein concentration. | SDS-PAGE enables you to visualize the presence of protein and provides information concerning the purity of your protein sample(s). Though comparing SDS-PAGE band intensities between samples and molecular weight standards gives an estimate of protein concentration, using a standard curve generated from samples of known protein concentration is a more precise method for measuring protein concentration. | ||
| − | <font color = #0d368e>'''To ensure the steps included below are clear, please watch the video tutorial linked here: [[https://www.dropbox.com/s/tzu2n1tqijeijk6/BCA.mp4?dl=0 BCA assay]]. | + | <font color = #0d368e>'''To ensure the steps included below are clear, please watch the video tutorial linked here: [[https://www.dropbox.com/s/tzu2n1tqijeijk6/BCA.mp4?dl=0 BCA assay]]. '''</font color> |
'''Prepare diluted bovine serum albumin (BSA) standards''' | '''Prepare diluted bovine serum albumin (BSA) standards''' | ||
| Line 145: | Line 124: | ||
'''Prepare Working Reagent (WR) and determine protein concentration''' | '''Prepare Working Reagent (WR) and determine protein concentration''' | ||
| − | #Use the following formula to calculate the volume of WR required: (# of standards + # unknowns) * 0.215 = total volume of WR (in mL). | + | #Use the following formula to calculate the volume of WR required: (# of standards + # unknowns) * 0.215 = total volume of WR (in mL). #Standards and unknowns will be run in duplicate. |
#Prepare the calculated volume of WR by mixing 50 parts of BCA Reagent A with 1 part BCA Reagent B (50:1 or 50-fold dilution of B). | #Prepare the calculated volume of WR by mixing 50 parts of BCA Reagent A with 1 part BCA Reagent B (50:1 or 50-fold dilution of B). | ||
#*For example, if your calculated total volume of WR is 100 mL, then mix 98 mL of A, and 2 mL of B. | #*For example, if your calculated total volume of WR is 100 mL, then mix 98 mL of A, and 2 mL of B. | ||
| − | #Pipet 25 μL of each standard prepared in the previous section and 25 μL of your concentrated | + | #Pipet 25 μL of each standard prepared in the previous section and 25 μL of your concentrated TDP43-RRM12 protein into separate, wells of a flat bottom 96-well plate. |
#Add 200 μL of the WR to each 25 μL aliquot of the standard and to the TDP43-RRM12 concentrated protein. | #Add 200 μL of the WR to each 25 μL aliquot of the standard and to the TDP43-RRM12 concentrated protein. | ||
#*The WR is added to each of the wells that was prepared in Step #3. | #*The WR is added to each of the wells that was prepared in Step #3. | ||
| Line 154: | Line 133: | ||
#Cool the plate to room temperature. | #Cool the plate to room temperature. | ||
#Using a plate reader spectrophotometer, measure the A<sub>562</sub> for each well. | #Using a plate reader spectrophotometer, measure the A<sub>562</sub> for each well. | ||
| − | |||
| − | |||
#Subtract the A<sub>562</sub> of the blank standard (I) from that of all the standard and TDP43-RRM12 samples. | #Subtract the A<sub>562</sub> of the blank standard (I) from that of all the standard and TDP43-RRM12 samples. | ||
#*This step subtracts the background 'noise' from the data measurements. | #*This step subtracts the background 'noise' from the data measurements. | ||
| Line 170: | Line 147: | ||
==Reagents list== | ==Reagents list== | ||
| − | * | + | *6x Reducing Laemmli Sample Buffer (from Boston BioProducts) |
*4-20% polyacrylamide gels in Tris-HCl (from Bio-Rad) | *4-20% polyacrylamide gels in Tris-HCl (from Bio-Rad) | ||
*TGS buffer: 5 mM Tris, 192 mM glycine, 0.1% (w/v) SDS (pH 8.3) (from Bio-Rad) | *TGS buffer: 5 mM Tris, 192 mM glycine, 0.1% (w/v) SDS (pH 8.3) (from Bio-Rad) | ||
| Line 177: | Line 154: | ||
*BioSafe Coomassie G-250 Stain (from Bio-Rad) | *BioSafe Coomassie G-250 Stain (from Bio-Rad) | ||
*Pierce BCA protein assay (from ThermoFisher) | *Pierce BCA protein assay (from ThermoFisher) | ||
| − | |||
==Navigation links== | ==Navigation links== | ||
Next day: [[20.109(S22):M1D5 |Perform aggregation assay using TDP43 protein and draft data slide for Research summary]] <br> | Next day: [[20.109(S22):M1D5 |Perform aggregation assay using TDP43 protein and draft data slide for Research summary]] <br> | ||
Previous day: [[20.109(S22):M1D3 |Induce and purify TDP43 protein]] <br> | Previous day: [[20.109(S22):M1D3 |Induce and purify TDP43 protein]] <br> | ||
Latest revision as of 20:13, 15 February 2022
Contents
Introduction
Two metrics will be used to evaluate the success of the protein purification protocol used to express and isolate TDP43-RRM12: purity and concentration.
Protein purity
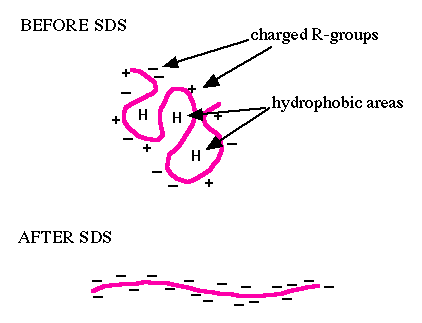
Electrophoresis is a technique that separates large molecules by size using an applied electrical field and a sieving matrix. DNA, RNA and proteins are the molecules most often studied with this technique; agarose and acrylamide gels are the two most common sieves. The molecules to be separated enter the matrix through a well at one end and are pulled through the matrix when a current is applied across it. The larger molecules get entwined in the matrix and are stalled; the smaller molecules wind through the matrix more easily and travel farther away from the well. The distance a nucleic acid or amino acid fragment travels is inversely proportional to the log of its length. Over time fragments of similar length accumulate into “bands” in the gel.
Protein concentration
To measure the concentration of your purified protein, you will use the BCA Protein Assay Reagent Kit. This kit enables colorimetric detection and quantification of the total protein within a sample. The ability to measure protein concentration is based on the detection of Cu1+ by the detection reagent, bicinchoninic acid (BCA). The Cu1+ is formed when Cu2+ is reduced by protein in an alkaline environment. Through this reduction reaction, a purple product is formed by the chelation of BCA and Cu1+ at a 2:1 ratio. This water-soluble complex has a strong absorbance at 562 nm. Because absorbance and protein concentration have a linear relationship, it is possible to compare the absorbance of an unknown protein sample to a standard curve, generated with samples of known protein concentrations, and calculate the concentration of protein in the experimental sample.
Protocols
Part 1: Participate in Communication Lab workshop
Our communication instructor, Dr. Prerna Bhargava, will join us today for a discussion on crafting data figures and legends.
Part 2: Visualize TDP43-RRM12 protein using polyacrylamide gel electrophoresis (PAGE)
As discussed above, SDS-PAGE is used to evaluate the purity protein. It is also possible to track the purification process using SDS-PAGE. For this, several aliquots were collected during the protein purification and concentration procedures. Each of these aliquots will be electrophoresed and the presence / absence of the protein of interest in each of the aliquots will provide information on the success of the purification.
One important note to consider is that the expected size of the TDP43-RRM12 product is dependent on the features that were added during the cloning process (review your notes from M2D1 prelab discussion!). The molecular weight of TDP43-RRM12 is ~20 kDa. Each of the features adds 'weight' to the product: SnapTag is ~20 kDa, 3C recognition site is ~1 kDa, MBP is ~40 kDa, and 6x-His is ~1 kDa. As you evaluate each lane of the SDS-PAGE results below, critically think about what the expected band size is for each of the samples. Review the M1D3 prelab slides for the orientation of the components that makeup the purified product.
To see a closer view of loading protein sample into an SDS-PAGE gel, please watch the video linked here: [Loading samples into a protein gel].
- Retrieve the aliquots that were collected during the protein purification and concentration procedures.
- In addition to the aliquots that were prepared in the previous laboratory session, you will also include the desalted protein in the SDS-PAGE experiment. To do this, transfer 30 μL from the 15 mL conical with your desalted protein into a microcentrifuge tube.
- To 30 μL of each aliquot, add 6 μL of Laemmli sample buffer.
- Prepare the cell pellet by adding 30 μL of water to a fresh microcentrifuge tube, then dip a clean pipette tip in the bacterial pellet and swirl into the water. Lastly, add 6 μL of Laemmli sample buffer.
- Boil all samples for 5 min in the water bath (set at 100 °C) located at the front laboratory bench.
- Secure the tubes with the cap-locks located next to baths to ensure that the caps do not pop open during the boiling step as this will result in your sample evaporating from the tube.
- Vortex and centrifuge samples for 15 seconds each to resuspend and collect samples in the bottoms of the tubes, respectively. Vortex and table-top mini centrifuge are at front bench.
- You will load all samples and a molecular weight standard.
- Load the total volume for the samples and 10 μL of the ladder into the polyacrylamide gel.
- Electrophorese at 200 V for 30-45 min.
- Following electrophoresis, remove the cartridge from the electrophoresis chamber and carefully pry apart the plastic plates that encase the polyacrylamide gel.
- With wet gloves, transfer the polyacrylamide gel to a staining box and add enough dH2O to cover the gel.
- Wash the gel for 5 min at room temperature on the rotating table.
- Decant the water from the staining box in the sink.
- Be careful that the gel does not fall into the sink!
- Repeat Steps #10-11 a total of 3 times.
- Cover the gel with the BioSafe Coomassie in the staining box and incubate on the rotating table at room temperature overnight.
- Empty the BioSafe Coomassie into the appropriate waste container in the chemical fume hood.
- Be careful that the gel does not fall into the waste container!
- Add 200 mL of dH2O to the staining box to wash the excess stain from the gel.
- Incubate on the rotating table at room temperature for 2 hrs.
- Replace the dH2O every 30 min.
- Image gel using the white light setting, not the UV light setting, of the gel documentation station.
In your laboratory notebook, complete the following:
- Consider the expected results for each lane in the polyacrylamide gel.
- Do you expect to see the TDP43-RRM12 protein in the lane? If yes, what is the expected size? If no, why not?
- Do you expect to see other cellular proteins in the lane? If yes, why? If no, why not?
Part 3: Measure protein concentration
SDS-PAGE enables you to visualize the presence of protein and provides information concerning the purity of your protein sample(s). Though comparing SDS-PAGE band intensities between samples and molecular weight standards gives an estimate of protein concentration, using a standard curve generated from samples of known protein concentration is a more precise method for measuring protein concentration.
To ensure the steps included below are clear, please watch the video tutorial linked here: [BCA assay].
Prepare diluted bovine serum albumin (BSA) standards
- Obtain a 1 mL aliquot of 2.0 mg/mL BSA stock.
- Prepare your standards according to the table below using PBS as the diluent:
| Tube |
Volume of diluent (μL) | Volume of BSA (μL) | Final concentration of BSA (μg/mL) |
|---|---|---|---|
| A | 0 | 300 of stock | 2000 |
| B | 125 | 375 of stock | 1500 |
| C | 325 | 325 of stock | 1000 |
| D | 175 | 175 of B | 750 |
| E | 325 | 325 of C | 500 |
| F | 325 | 325 of E | 250 |
| G | 325 | 325 of F | 125 |
| H | 400 | 100 of G | 25 |
| I | 400 | 0 | 0 = blank |
Prepare Working Reagent (WR) and determine protein concentration
- Use the following formula to calculate the volume of WR required: (# of standards + # unknowns) * 0.215 = total volume of WR (in mL). #Standards and unknowns will be run in duplicate.
- Prepare the calculated volume of WR by mixing 50 parts of BCA Reagent A with 1 part BCA Reagent B (50:1 or 50-fold dilution of B).
- For example, if your calculated total volume of WR is 100 mL, then mix 98 mL of A, and 2 mL of B.
- Pipet 25 μL of each standard prepared in the previous section and 25 μL of your concentrated TDP43-RRM12 protein into separate, wells of a flat bottom 96-well plate.
- Add 200 μL of the WR to each 25 μL aliquot of the standard and to the TDP43-RRM12 concentrated protein.
- The WR is added to each of the wells that was prepared in Step #3.
- Cover your plate with plastic wrap and incubate at 37 °C incubator for 30 min.
- Cool the plate to room temperature.
- Using a plate reader spectrophotometer, measure the A562 for each well.
- Subtract the A562 of the blank standard (I) from that of all the standard and TDP43-RRM12 samples.
- This step subtracts the background 'noise' from the data measurements.
- Generate a standard curve by plotting the blank-corrected A562 for each BSA standard (A-I, B-I, ..., I-I) vs. its concentration in μg/mL.
- Use the standard curve to calculate the concentration of protein in the TDP43-RRM12 sample.
In your laboratory notebook, complete the following:
- Graph a standard curve using the data collected from the BCA assay.
- Include the equation of the trend line and R2 value.
- What does the R2 indicate about the data that there were collected? Are you confident in these data? Why or why not?
- Use the equation of the trendline to calculate the concentration of TDP43-RRM12 in protein solution.
- Why is it useful to evaluate the purity and measure the concentration of protein that will be used in experiments?
- Hint: consider the specificity of each technique and what information is provided by each method. Is TDP43-RRM12 the only protein present in the concentrated sample? How do you know?
Reagents list
- 6x Reducing Laemmli Sample Buffer (from Boston BioProducts)
- 4-20% polyacrylamide gels in Tris-HCl (from Bio-Rad)
- TGS buffer: 5 mM Tris, 192 mM glycine, 0.1% (w/v) SDS (pH 8.3) (from Bio-Rad)
- Precision Plus Dual Color Standard ladder (from Bio-Rad)
- Molecular weights of ladder bands (linked here).
- BioSafe Coomassie G-250 Stain (from Bio-Rad)
- Pierce BCA protein assay (from ThermoFisher)
Next day: Perform aggregation assay using TDP43 protein and draft data slide for Research summary