Difference between revisions of "20.109(S18):Induce DNA damage and apply drug treatments for RNA purification (Day2)"
Noreen Lyell (Talk | contribs) (→Part 2: Research tissue culture cell lines) |
(→Part 1: Induce DNA damage) |
||
| (34 intermediate revisions by 2 users not shown) | |||
| Line 4: | Line 4: | ||
==Introduction== | ==Introduction== | ||
| − | In this module, you will assess gene expression using two methods: quantitative PCR (qPCR) and RNA-seq. For timing reasons you will not be able to prepare and submit your treated samples for RNA-seq. Instead you will use RNA-seq data generated by the teaching faculty. You will, however, complete the same procedure used by the teaching faculty to prepare the RNA samples and then use your samples in a qPCR experiment to confirm the RNA-seq results. | + | In this module, you will assess gene expression using two methods: quantitative PCR (qPCR) and RNA-seq. For timing reasons you will not be able to prepare and submit your treated samples for RNA-seq. Instead you will use RNA-seq data generated by the teaching faculty. You will, however, complete the same procedure used by the teaching faculty to prepare the RNA samples and then use your samples in a qPCR experiment to confirm the RNA-seq results. |
| − | + | To begin your assay, you will first induce DNA damage by incubating your cells with etoposide. Etoposide is itself a chemotherapy drug that generates DNA stand breaks by forming a ternary complex with DNA and topoisomerase II. This prevents religation of the DNA strands after unwinding. This effects cancer cells more than non-cancerous cells because cancer cells divide much faster. We will use etoposide to damage the DNA then examine the differences in gene expression of DLD-1 compared to BRCA2-/- cells. | |
| − | + | To check that DNA damage was induced by the etoposide treatment, you will measure gene expression of a DNA cell cycle checkpoint factor using quantitative PCR (qPCR). In qPCR, the amount of a specific transcript can be measured. The presence of checkpoint factors in an indication of DNA damage in that the cell cycle is halted when excessive damage is incurred by a cell. For your experiments, the results of p21 expression levels will serve as a control that DNA damage was indeed induced following etoposide treatment. In addition, the qPCR results will be compared to the expression levels measured via RNA-seq. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
==Protocols== | ==Protocols== | ||
| − | ===Part 1: Induce DNA damage | + | ===Part 1: Induce DNA damage=== |
| − | + | In this exercise, you will induce DNA damage in cells that were seeded by the teaching faculty (using the cells you seeded during the previous laboratory session). Next class you will purify RNA from your cells for quantitative PCR analysis. Control, no treatment flasks will be taken care of by instructors. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
[[Image:Sp18 20.109 M2 qPCR overview.png|thumb|750px|center|]] | [[Image:Sp18 20.109 M2 qPCR overview.png|thumb|750px|center|]] | ||
| − | |||
#Prepare your working space within the tissue culture hood. | #Prepare your working space within the tissue culture hood. | ||
#Calculate the volume of etoposide stock needed for DNA damage induction. | #Calculate the volume of etoposide stock needed for DNA damage induction. | ||
| − | #*Obtain an aliquot of pre-warmed media from the 37 °C water bath | + | #*Obtain an aliquot of pre-warmed media from the 37 °C water bath. |
| − | #*Determine the volume of etoposide stock (100 mM) you need to add to the media for a final concentration of 100 μM. | + | #*Transfer 10 mL of the media into a 15 mL conical tube. |
| − | #Retrieve | + | #*Determine the volume of etoposide stock (100 mM) you need to add to the media for a final concentration of 100 μM in the 10 mL aliquot. |
| + | #*Prepare your media +etoposide. | ||
| + | #Retrieve two T75 flasks (one DLD-1 culture and one BRCA2-/- culture) from the 37 °C incubator and visually inspect your cells with a microscope. | ||
#*Record your observations concerning media color, confluency, etc. in your laboratory notebook. | #*Record your observations concerning media color, confluency, etc. in your laboratory notebook. | ||
#Move your flasks into the tissue culture hood. | #Move your flasks into the tissue culture hood. | ||
| Line 96: | Line 30: | ||
#Aspirate the PBS from each flask. | #Aspirate the PBS from each flask. | ||
#*Again, be careful not to cross-contaminate. | #*Again, be careful not to cross-contaminate. | ||
| − | #Add 5 mL of the media containing etoposide that you prepared in Step #2. | + | #Add 5 mL of the media containing etoposide that you prepared in Step #2 to the flasks. |
| − | #Carefully | + | #Carefully move the 'etoposide treated' flasks to the 37 °C incubator for 60 min. |
| − | #Assist | + | #Assist your partner if they are still working. When you are both done, return to the main laboratory space. |
#Return to the tissue culture space and retrieve your flasks from the 37 °C incubator. | #Return to the tissue culture space and retrieve your flasks from the 37 °C incubator. | ||
#Aspirate the media containing etoposide from each flask. | #Aspirate the media containing etoposide from each flask. | ||
#*Be mindful of cross-contamination. | #*Be mindful of cross-contamination. | ||
#Add 10 mL of fresh media to each flask. | #Add 10 mL of fresh media to each flask. | ||
| − | #Label | + | #Label the flasks containing the DLD-1 and one BRCA2-/- cultures that were treated with etoposide with '+etop' to denote that the cells were treated with only etoposide. |
| − | #* | + | #*Move these flasks to the 37 °C incubator. |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
#Assist your laboratory partner, if necessary, then clean the hood and return the main laboratory space. | #Assist your laboratory partner, if necessary, then clean the hood and return the main laboratory space. | ||
===Part 2: Research tissue culture cell lines=== | ===Part 2: Research tissue culture cell lines=== | ||
| − | In the previous laboratory session, you practiced tissue culture techniques to split, or culture, the cell lines that you will use in this module. Specifically, you will assess gene expression in DLD-1 parental and BRCA2- mutant cells. In this exercise, you will learn more about these cell lines to prepare you for your experiments and your M2 Research article. | + | In the previous laboratory session, you practiced tissue culture techniques to split, or culture, the cell lines that you will use in this module. Specifically, you will assess gene expression in DLD-1 parental and BRCA2-/- mutant cells. In this exercise, you will learn more about these cell lines to prepare you for your experiments and your M2 Research article. |
| − | The parental strain is DLD-1, and an important resource for learning about common cell lines is the American Type Culture Collection, or ATCC. Visit the | + | The parental strain is DLD-1, and an important resource for learning about common cell lines is the American Type Culture Collection, or ATCC. Visit the [https://www.atcc.org/products/all/CCL-221.aspx#generalinformation DLD-1 homepage] and answer the questions below in your laboratory notebook. |
| − | #From what source were these | + | #From what source were these cells derived (organism and tissue)? What other details concerning the host are provided? |
#Do these cells float in the culture media, or stick to the culture dish? How do you know? | #Do these cells float in the culture media, or stick to the culture dish? How do you know? | ||
#What is the morphology of these cells? What does this mean? | #What is the morphology of these cells? What does this mean? | ||
| − | #Carefully read through the Culture Methods | + | #Carefully read through the Culture Methods provided for these cells. Does this procedure differ from what you did in the previous laboratory session? How? |
The BRCA2-/- mutant strain was generated via targeted disruption of BRCA2 exon 11 using recombinant adeno-associated virus gene editing technology. DLD-1 and BRCA2-/- are isogenic cell lines. | The BRCA2-/- mutant strain was generated via targeted disruption of BRCA2 exon 11 using recombinant adeno-associated virus gene editing technology. DLD-1 and BRCA2-/- are isogenic cell lines. | ||
| Line 126: | Line 54: | ||
#What does it mean when cell lines are isogenic? Why is this useful in research? | #What does it mean when cell lines are isogenic? Why is this useful in research? | ||
| − | Lastly, to provide context for your research you will review the following journal article concerning BRCA2- | + | Lastly, to provide context for your research you will review the following journal article concerning the BRCA2-/- cell line and answer the questions below. |
| − | [[Media: | + | *[[Media:BRCA2 reference, Hucl et al.pdf| A syngeneic variance library for functional annotation of human variation: application to BRCA2.]] (2008) ''Cancer Res'' by Hucl et al. |
| + | #Read the Introduction. Given the information provided here and in lecture, why was exon 11 targeted to generate the BRCA2-/2 mutant (ie why does this result in a BRCA2 null mutant)? | ||
| + | #Review Figure 2 and the following results sub-sections: RAD51 focus formation, Chromosomal instability, Cell survival after irradiation and Cell proliferation on drug treatment. | ||
| + | #*The authors report that RAD51 focus formation and chromosomal aberrations are decreased and increased, respectively in BRCA2-/- cells compared to DLD-1 cells after treatment with a DNA-damaging agent. What are the implications of this in disease, specifically why might these outcomes result in higher rates of cancer in BRCA2 null individuals? | ||
| + | #*Cell survival following irradiation and various drug treatments are reported in the results. Given the data, which treatment is better suited to treat patients with BRCA2 null cancer? Use what you know about the role of BRCA2 and the type of DNA damage induced by the treatment agent to support your answer. | ||
| + | #Write a hypothesis that states your expectation(s) related to cell survival for the conditions you will test: DLD-1 and BRCA2-/- with no treatment, with etoposide, and with etoposide plus increasing drug concentration. | ||
==Reagents== | ==Reagents== | ||
*etoposide, stock = 100 mM (Sigma-Aldrich) | *etoposide, stock = 100 mM (Sigma-Aldrich) | ||
| − | |||
| − | |||
==Navigation links== | ==Navigation links== | ||
Next day: [[20.109(S18):Purify RNA and practice RNA-seq data analysis methods (Day3)| Purify RNA and practice RNA-seq data analysis methods]] | Next day: [[20.109(S18):Purify RNA and practice RNA-seq data analysis methods (Day3)| Purify RNA and practice RNA-seq data analysis methods]] | ||
Previous day: [[20.109(S18):Practice tissue culture techniques and prepare cells for RNA purification (Day1)| Practice tissue culture techniques and prepare cells for RNA purification]] | Previous day: [[20.109(S18):Practice tissue culture techniques and prepare cells for RNA purification (Day1)| Practice tissue culture techniques and prepare cells for RNA purification]] | ||
Latest revision as of 14:24, 14 March 2018
Contents
Introduction
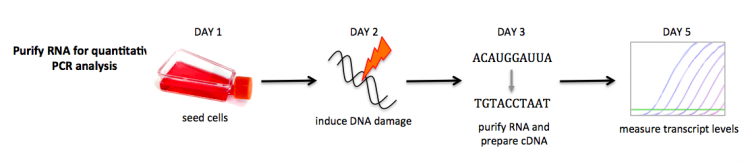
In this module, you will assess gene expression using two methods: quantitative PCR (qPCR) and RNA-seq. For timing reasons you will not be able to prepare and submit your treated samples for RNA-seq. Instead you will use RNA-seq data generated by the teaching faculty. You will, however, complete the same procedure used by the teaching faculty to prepare the RNA samples and then use your samples in a qPCR experiment to confirm the RNA-seq results.
To begin your assay, you will first induce DNA damage by incubating your cells with etoposide. Etoposide is itself a chemotherapy drug that generates DNA stand breaks by forming a ternary complex with DNA and topoisomerase II. This prevents religation of the DNA strands after unwinding. This effects cancer cells more than non-cancerous cells because cancer cells divide much faster. We will use etoposide to damage the DNA then examine the differences in gene expression of DLD-1 compared to BRCA2-/- cells.
To check that DNA damage was induced by the etoposide treatment, you will measure gene expression of a DNA cell cycle checkpoint factor using quantitative PCR (qPCR). In qPCR, the amount of a specific transcript can be measured. The presence of checkpoint factors in an indication of DNA damage in that the cell cycle is halted when excessive damage is incurred by a cell. For your experiments, the results of p21 expression levels will serve as a control that DNA damage was indeed induced following etoposide treatment. In addition, the qPCR results will be compared to the expression levels measured via RNA-seq.
Protocols
Part 1: Induce DNA damage
In this exercise, you will induce DNA damage in cells that were seeded by the teaching faculty (using the cells you seeded during the previous laboratory session). Next class you will purify RNA from your cells for quantitative PCR analysis. Control, no treatment flasks will be taken care of by instructors.
- Prepare your working space within the tissue culture hood.
- Calculate the volume of etoposide stock needed for DNA damage induction.
- Obtain an aliquot of pre-warmed media from the 37 °C water bath.
- Transfer 10 mL of the media into a 15 mL conical tube.
- Determine the volume of etoposide stock (100 mM) you need to add to the media for a final concentration of 100 μM in the 10 mL aliquot.
- Prepare your media +etoposide.
- Retrieve two T75 flasks (one DLD-1 culture and one BRCA2-/- culture) from the 37 °C incubator and visually inspect your cells with a microscope.
- Record your observations concerning media color, confluency, etc. in your laboratory notebook.
- Move your flasks into the tissue culture hood.
- Aspirate the spent media from each flask.
- Be careful not to cross-contaminant between flasks with different cell lines!
- Add 5 mL of PBS to each flask and rock the plate gently to wash the cells.
- Aspirate the PBS from each flask.
- Again, be careful not to cross-contaminate.
- Add 5 mL of the media containing etoposide that you prepared in Step #2 to the flasks.
- Carefully move the 'etoposide treated' flasks to the 37 °C incubator for 60 min.
- Assist your partner if they are still working. When you are both done, return to the main laboratory space.
- Return to the tissue culture space and retrieve your flasks from the 37 °C incubator.
- Aspirate the media containing etoposide from each flask.
- Be mindful of cross-contamination.
- Add 10 mL of fresh media to each flask.
- Label the flasks containing the DLD-1 and one BRCA2-/- cultures that were treated with etoposide with '+etop' to denote that the cells were treated with only etoposide.
- Move these flasks to the 37 °C incubator.
- Assist your laboratory partner, if necessary, then clean the hood and return the main laboratory space.
Part 2: Research tissue culture cell lines
In the previous laboratory session, you practiced tissue culture techniques to split, or culture, the cell lines that you will use in this module. Specifically, you will assess gene expression in DLD-1 parental and BRCA2-/- mutant cells. In this exercise, you will learn more about these cell lines to prepare you for your experiments and your M2 Research article.
The parental strain is DLD-1, and an important resource for learning about common cell lines is the American Type Culture Collection, or ATCC. Visit the DLD-1 homepage and answer the questions below in your laboratory notebook.
- From what source were these cells derived (organism and tissue)? What other details concerning the host are provided?
- Do these cells float in the culture media, or stick to the culture dish? How do you know?
- What is the morphology of these cells? What does this mean?
- Carefully read through the Culture Methods provided for these cells. Does this procedure differ from what you did in the previous laboratory session? How?
The BRCA2-/- mutant strain was generated via targeted disruption of BRCA2 exon 11 using recombinant adeno-associated virus gene editing technology. DLD-1 and BRCA2-/- are isogenic cell lines.
- Provide a brief description of how adeno-associated virus gene editing technology disrupts genes to generate mutant cell lines.
- What does it mean when cell lines are isogenic? Why is this useful in research?
Lastly, to provide context for your research you will review the following journal article concerning the BRCA2-/- cell line and answer the questions below.
- A syngeneic variance library for functional annotation of human variation: application to BRCA2. (2008) Cancer Res by Hucl et al.
- Read the Introduction. Given the information provided here and in lecture, why was exon 11 targeted to generate the BRCA2-/2 mutant (ie why does this result in a BRCA2 null mutant)?
- Review Figure 2 and the following results sub-sections: RAD51 focus formation, Chromosomal instability, Cell survival after irradiation and Cell proliferation on drug treatment.
- The authors report that RAD51 focus formation and chromosomal aberrations are decreased and increased, respectively in BRCA2-/- cells compared to DLD-1 cells after treatment with a DNA-damaging agent. What are the implications of this in disease, specifically why might these outcomes result in higher rates of cancer in BRCA2 null individuals?
- Cell survival following irradiation and various drug treatments are reported in the results. Given the data, which treatment is better suited to treat patients with BRCA2 null cancer? Use what you know about the role of BRCA2 and the type of DNA damage induced by the treatment agent to support your answer.
- Write a hypothesis that states your expectation(s) related to cell survival for the conditions you will test: DLD-1 and BRCA2-/- with no treatment, with etoposide, and with etoposide plus increasing drug concentration.
Reagents
- etoposide, stock = 100 mM (Sigma-Aldrich)
Next day: Purify RNA and practice RNA-seq data analysis methods
Previous day: Practice tissue culture techniques and prepare cells for RNA purification