20.109(S18):Analyze RNA-seq data and prepare for quantitative PCR experiment (Day 4)
Contents
Introduction
The transcriptome is the full suite of transcripts within an organism and provides the key link between the genetic code and phenotype. Research focused on the transcriptome has provided important insights into how gene expression is altered in different cell / tissue types, in developmental phases, in disease states, and between species. In this module, you will evaluate gene expression differences in the parental DLD-1 cell line compared to the BRCA2-/2 mutant cell line. In addition, you will assess the effects of DNA damage and drug treatments on the transcriptome in these cells.
The gene expression data was generated using RNA-seq. In this method deep sequencing is completed using reagents and equipment from Illumina. With this technology, transcripts are directly sequenced and mapped to a reference genome. Then the reads are counted to provide information on gene expression levels for a particular portion of the genome (i.e. for a particular gene).
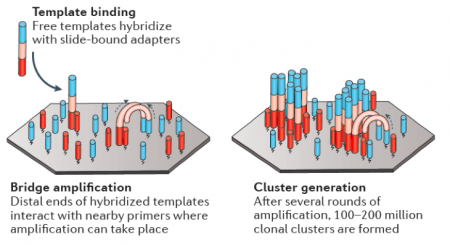
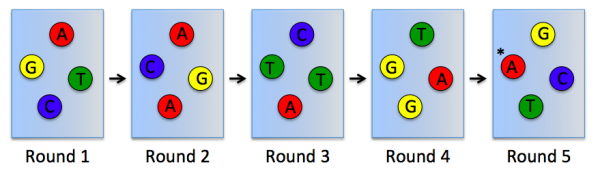
In RNA-seq, RNA is purified from cells and reverse transcribed into DNA. The DNA molecules are modified with adapters, which are ligated to both ends of the DNA. Sequences complementary to the adapters are attached to the surface of flow cell channels and facilitate binding of modified DNA molecules and provide a primer for DNA polymerase. Following the initial binding to the flow cell channel, the DNA molecules from bridges that enable bridge amplification and cluster generation (see figure to the right). Through this process millions of dense clusters containing double-stranded DNA are generated.To directly sequence from the clusters a sequencing by synthesis approach is used. In this, several rounds of amplification are performed using deoxynucleoside triphosphate (dNTP) bases. dNTP are terminator molecules given that the ribose 3'-OH group is blocked thereby preventing elongation by polymerase. Each terminating base (dATP, dTTP, dCTP, and dGTP) is fluorescently labeled (dATP = red, dTTP = green, dCTP = blue, and dGTP = yellow). For each round of sequencing a mixture containing all four labelled dNTP bases is added and a single base is incorporated to each DNA molecule bound to the flow cell channels. The flow cell is then imaged to capture the dNTP base that was added at each cluster location. Then the fluorescent label and 3'-OH blocking group is removed from the incorporated dNTP and another round of sequencing is performed. This results in the full sequences of every DNA molecule bound to the flow cell channels. Therefore, the sequence of the cluster denoted by the asterisk is GCTGA in the schematic provided below.
As with all technologies, there are positives and negatives to RNA-seq. On the plus side, the ability to directly sequence enables researchers to assess gene expression in organisms for which a full genome sequence is not available or not fully annotated. Furthermore, this method allows for the quantification of individual isoforms that result from alternate splicing. On this minus side, the cost of RNA-seq can limit the depth of sequencing achieved and genes that are not highly expressed may not be captured in a data set.
Protocols
Part 1: Analyze RNA-seq data
Today you will analyze the RNA-seq data gathered from untreated DLD-1 and BRCA2- cells and etoposide treated DLD-1 and BRCA2- cells. Following RNA purification, the samples were submitted to the BioMicro Center for Illumina sequencing. Illumina sequencing technology, or sequencing by synthesis (SBS), is used for massively parallel sequencing with a proprietary method that detects single bases as they are incorporated into growing DNA strands.
Analysis of RNA-Seq data by Amanda Kedaigle, Prof. Ernest Fraenkel & Prof. Leona Samson.
Part 2: Design primers for qPCR
Next day: Investigate RNA-seq data using public databases
Previous day: Purify RNA and practice RNA-seq data analysis methods