Difference between revisions of "20.109(S18):Analyze RNA-seq data and prepare for quantitative PCR experiment (Day 4)"
Noreen Lyell (Talk | contribs) (→Introduction) |
Noreen Lyell (Talk | contribs) (→Protocols) |
||
| Line 16: | Line 16: | ||
==Protocols== | ==Protocols== | ||
| + | |||
| + | ===Part 1: Analyze RNA-seq data=== | ||
Today you will analyze the RNA-seq data gathered from untreated DLD-1 and BRCA2- cells and etoposide treated DLD-1 and BRCA2- cells. Following RNA purification, the samples were submitted to the BioMicro Center for Illumina sequencing. Illumina sequencing technology, or sequencing by synthesis (SBS), is used for massively parallel sequencing with a proprietary method that detects single bases as they are incorporated into growing DNA strands. | Today you will analyze the RNA-seq data gathered from untreated DLD-1 and BRCA2- cells and etoposide treated DLD-1 and BRCA2- cells. Following RNA purification, the samples were submitted to the BioMicro Center for Illumina sequencing. Illumina sequencing technology, or sequencing by synthesis (SBS), is used for massively parallel sequencing with a proprietary method that detects single bases as they are incorporated into growing DNA strands. | ||
[[ Analysis of RNA-Seq data (Day 4)| Analysis of RNA-Seq data]] by Amanda Kedaigle, Prof. Ernest Fraenkel & Prof. Leona Samson.<br> | [[ Analysis of RNA-Seq data (Day 4)| Analysis of RNA-Seq data]] by Amanda Kedaigle, Prof. Ernest Fraenkel & Prof. Leona Samson.<br> | ||
| + | |||
| + | ===Part 2: Design primers for qPCR=== | ||
==Navigation links== | ==Navigation links== | ||
Next day: [[20.109(S18):Investigate RNA-seq data using public databases (Day5) | Investigate RNA-seq data using public databases]] | Next day: [[20.109(S18):Investigate RNA-seq data using public databases (Day5) | Investigate RNA-seq data using public databases]] | ||
Previous day: [[20.109(S18):Purify RNA and practice RNA-seq data analysis methods (Day3)| Purify RNA and practice RNA-seq data analysis methods]] | Previous day: [[20.109(S18):Purify RNA and practice RNA-seq data analysis methods (Day3)| Purify RNA and practice RNA-seq data analysis methods]] | ||
Revision as of 19:35, 5 January 2018
Contents
Introduction
Quantitative polymerase chain reaction (qPCR) allows researchers to monitor the results of PCR as amplification is occurring (this technique is also referred to as real-time polymerase chain reaction or real-time PCR). During qPCR data are collected throughout the amplification process using a fluorescent dye. The fluorescent dye is highly specific for double-stranded DNA and when bound to DNA molecules the fluorescence intensity increases proportionately to the increase in double-stranded product. In contrast, the data for traditional PCR are simply observed as a band on a gel.
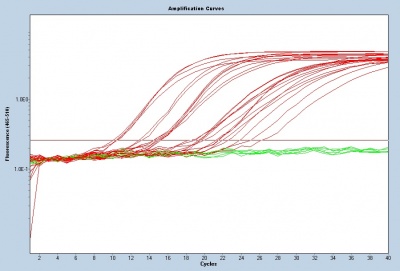
To assess gene transcript levels, you will examine the CT values from your qPCR assay. The CT values are displayed as an amplification curve following qPCR (these values are also given numerically). The initial cycles measure very little fluorescence due to low amounts of double-stranded DNA and are used to establish the inherent background fluorescence. As double-stranded product is produced, fluorescence is measured and the curve appears linear. This linear portion of the curve represents the exponential phase of PCR. Throughout the exponential phase, the curve should be smooth. Sharp points may be due to errors in reaction preparation or failures in the machine used to measure fluorescence. As mentioned previously, the first cycle in which the fluorescence measurement is above background is the CT. During the later cycles the curve shows minimal increases in fluorescence due the depletion of reagents.
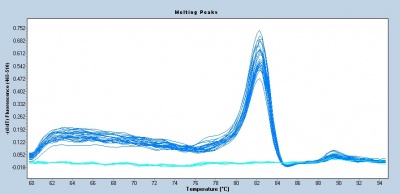
Following the qPCR amplification measurements, a melt curve is completed. Melt curves assess the dissociation of double-stranded DNA while the sample is heated. As the temperature is increased, double-stranded DNA ‘melts’ as the strands dissociate. As discussed above, the fluorescent dye used in qPCR associates with double-stranded DNA and fluorescence measurements will decrease as the temperature increases. In qPCR, the melt curve is used to confirm that a single amplification product was generated during the reaction. If additional products were present, the melt curve would presumably show additional peaks. Why might this be true? Can you think of a scenario where two different products would produce a single peak in a melt curve?
Protocols
Part 1: Analyze RNA-seq data
Today you will analyze the RNA-seq data gathered from untreated DLD-1 and BRCA2- cells and etoposide treated DLD-1 and BRCA2- cells. Following RNA purification, the samples were submitted to the BioMicro Center for Illumina sequencing. Illumina sequencing technology, or sequencing by synthesis (SBS), is used for massively parallel sequencing with a proprietary method that detects single bases as they are incorporated into growing DNA strands.
Analysis of RNA-Seq data by Amanda Kedaigle, Prof. Ernest Fraenkel & Prof. Leona Samson.
Part 2: Design primers for qPCR
Next day: Investigate RNA-seq data using public databases
Previous day: Purify RNA and practice RNA-seq data analysis methods