Difference between revisions of "20.109(S17):Module 1"
From Course Wiki
Noreen Lyell (Talk | contribs) |
Noreen Lyell (Talk | contribs) |
||
| Line 1: | Line 1: | ||
{{Template:20.109(S17)}} | {{Template:20.109(S17)}} | ||
<div style="padding: 10px; width: 790px; border: 5px solid #6633CC;"> | <div style="padding: 10px; width: 790px; border: 5px solid #6633CC;"> | ||
| + | |||
| + | =<center>Module 1</center>= | ||
| + | |||
| + | '''Lecturer:''' [http://be.mit.edu/directory/angela-koehler Angela Koehler]<br> | ||
| + | '''Instructors:''' [http://be.mit.edu/directory/noreen-lyell Noreen Lyell], [http://be.mit.edu/directory/leslie-mcclain Leslie McClain] and [http://be.mit.edu/directory/maxine-jonas Maxine Jonas] | ||
| + | |||
| + | '''TA:''' Rob Wilson <br> | ||
| + | '''Lab manager:''' Hsinhwa Lee <br> | ||
| + | |||
| + | ==Overview== | ||
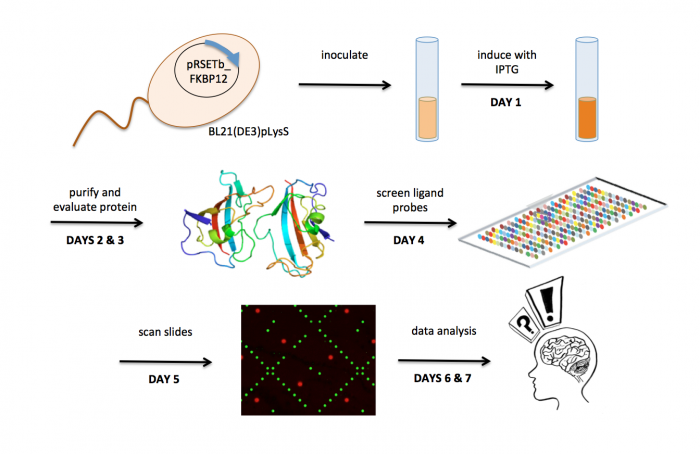
[[Image:Sp17 20.109 M1 overview.png|thumb|700px|center|]] | [[Image:Sp17 20.109 M1 overview.png|thumb|700px|center|]] | ||
| + | |||
| + | <br style="clear:both" /> | ||
| + | |||
| + | ==Lab links: day by day== | ||
| + | [[20.109(S17):In silico cloning and induction of protein expression (Day1) |In silico cloning and induce protein expression]]<br> | ||
| + | [[20.109(S17):Purification of induced protein (Day2) |Purify induced protein]]<br> | ||
| + | [[20.109(S17):Evaluation of purified protein (Day3) |Evaluate purity and concentration of protein]]y]]<br> | ||
| + | [[20.109(S17):Screen chemical library for FKBP12 binders (Day4) |Screen ligand library for FKBP12 binders]]<br> | ||
| + | [[20.109(S17):Scan slides to identify FKBP12 binders (Day5) |Scan slides to identify FKBP12 binders]]<br> | ||
| + | [[20.109(S17):Complete data analysis (Day6) |Complete data analysis]]<br> | ||
| + | [[20.109(S17):Identify chemical structures common among FKBP12 binders |Identify chemical structures common among FKBP12 binders]]<br> | ||
| + | |||
| + | ==Data== | ||
| + | See all M1 student data on the [http://engineerbiology.org/wiki/Talk:20.109(S17):Module_1 Discussion page]. | ||
| + | |||
| + | ==Assignments== | ||
| + | |||
| + | [[20.109(S17): M1 Data Summary | Data summary]] <br> | ||
| + | [[20.109(S17): M1 Mini-presentation | Mini-presentation]] <br> | ||
| + | |||
| + | ==References== | ||
| + | |||
| + | [[Media:Sp17 M1 reference NatProt.pdf| A method for the covalent capture and screening of diverse small molecules in a microarray format.]] ''Nature Protocols.'' 1:2344-2352. | ||
| + | |||
| + | [[Media:Sp17 M1 reference ChemBiol.pdf| Recent discoveries and applications involving small-molecule microarrays.]] ''Chemical Biology.'' 18:21-28. | ||
| + | |||
| + | ==Notes for teaching faculty== | ||
| + | [[20.109(S17): TA notes for M1| S17 notes for M1]] | ||
| + | [[20.109(S17): TA notes for orientation| S17 notes for orientation day]] | ||
Revision as of 22:46, 30 January 2017
Contents
Module 1
Lecturer: Angela Koehler
Instructors: Noreen Lyell, Leslie McClain and Maxine Jonas
TA: Rob Wilson
Lab manager: Hsinhwa Lee
Overview
Lab links: day by day
In silico cloning and induce protein expression
Purify induced protein
Evaluate purity and concentration of proteiny]]
Screen ligand library for FKBP12 binders
Scan slides to identify FKBP12 binders
Complete data analysis
Identify chemical structures common among FKBP12 binders
Data
See all M1 student data on the Discussion page.
Assignments
Data summary
Mini-presentation
References
A method for the covalent capture and screening of diverse small molecules in a microarray format. Nature Protocols. 1:2344-2352.
Recent discoveries and applications involving small-molecule microarrays. Chemical Biology. 18:21-28.