20.109(S17):Identify chemical structures common among FKBP12 binders
Contents
Introduction
Proteins and small molecules interact in a process referred to as molecular recognition. In molecular recognition, the non-covalent complexes that form are defined by two characteristics: specificity and affinity.
- Specificity distinguishes a specific binding partner from the milieu of potential binding partners in complex environments.
- Affinity dictates the likelihood of binding based on the concentration of a specific binding partner in a milieu of potential binding partners such that high affinity partners at low concentrations are not outcompeted by low affinity partners and high concentrations.
In cells, proteins are critical macromolecules that perform numerous roles related to structure, mechanics, metabolism, and signaling. In these roles, proteins do not work in a vacuum; rather the biological roles of proteins are dependent on direct physical interactions with other molecules. Though our focus in on small molecule binders, proteins also interact with other proteins, nucleic acids, oxygen, and metals. In all cases, the interactions are characterized by protein-ligand-solvent binding kinetics.
Though beyond the scope of this module, protein-ligand-solvent binding kinetics are defined as a thermodynamic system composed of solute (proteins and binders) and solvent (liquid that contains the proteins and binders). In that protein – small molecule complexes result in heat transfer, the driving forces that promote these interactions are due to energy exchanges that are characterized by Gibbs free energy (ΔG). ΔG measures the capacity of a thermodynamic system to do maximum or reversible work at constant temperature and pressure. When at equilibrium with constant temperature and pressue, protein – small molecule binding occurs when the change in ΔG is negative. The magnitude of ΔG provides insight into the stability of a protein – small molecule complex.
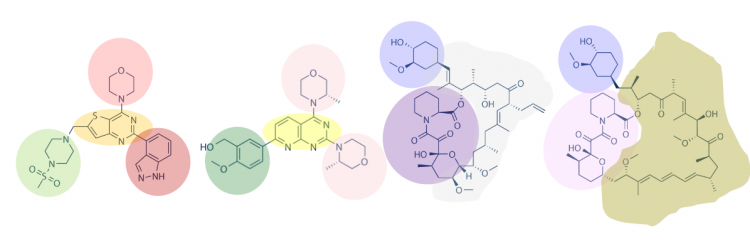
Another method for assessing protein – small molecule binding is to visually inspect known small molecule binders for common features / structures. Though you may not realize it, you did this on M1D2! Remember the information regarding lactose and IPTG? Or histidine and imidazole? In both of these examples, the molecules share a feature that enable binding to the same target (LacI for lactose and IPTG and Ni2+ for histidine and imidazole). Your goal for today is to carefully examine the hits identified by the class and identify any common features / structures. As in the image below, it is possible that multiple features will be present within the same small molecule.
Protocols
Part 1: Visually evaluate chemical structures of positive hits
If you haven't already uploaded the chemical structures for your top five hits to the Discussion tab associated with the M1 overview page, please do so now as your classmates will need your results for this exercise.
With your partner, visually inspect the hits identified in our SMM. It may be easier to print the small molecule images so you can readily see all of the structures. Also, it may be helpful to use a color-coding system (like the one in the image provided in the Introduction) to highlight features / structures that are common to the FKBP12 binders.
Discuss the following questions with your partner and record your thoughts in your laboratory notebook:
- How many common features did you identify? Are there more or less than you expected?
Part 2: Develop future works and implications section
Next day: First day of M2
Previous day: Complete data analysis