Difference between revisions of "20.109(S16):Introduction to cell strains and plating (Day1)"
MAXINE JONAS (Talk | contribs) (→Reagent list) |
(→Reagent list) |
||
| (54 intermediate revisions by 3 users not shown) | |||
| Line 5: | Line 5: | ||
==Introduction== | ==Introduction== | ||
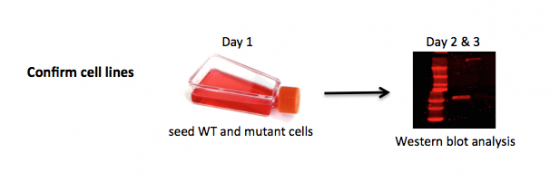
| − | + | To be confident of your findings related to DNA repair, you will first validate the cell lines you are using to assess the effect of DNA damage and drugs on NHEJ. Just as it was important to confirm the template you used for your mutagenesis reaction in Mod 1, it is important to confirm the cells you are using are indeed mutants in the protein you are assessing in this module. You will do this by completing a Western blot. A Western blot allows researchers to detect specific proteins within a mixed sample, such as cell lysate. We will use this protein analysis technique to assess DNA-PK levels in the wild-type M059K and DNA-PK mutant M059J cell lines. | |
| − | + | [[Image:Sp16 M2D1 cell confirmation.png|thumb|550px|center|'''Strategy for assessing DNA-PK levels in cell lines.''']] | |
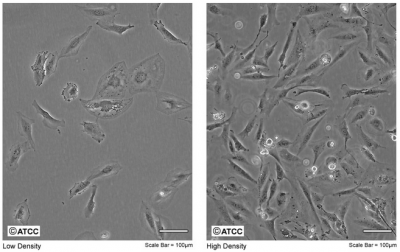
| − | + | [[Image:Sp16 M2D1 M059K cells.png|thumb|400px|right|'''M059K cells.''' The fibroblasts are shown at low (left) and high (right) cell densities.]]One important objective for this experimental module is for you to learn how to maintain cultured mammalian cell lines, or to perform tissue culture. Today you will jump right into this task as you seed M059K and M059J cells for your Western protein assay. These human fibroblast cell lines were originally acquired from a malignant glioblastoma. | |
| − | + | Tissue culture was developed ~100 years ago as a means to study mammalian biology, and since that time we have learned a tremendous amount by observing the behavior of mammalian cells maintained in the laboratory. The term tissue culture was originally coined because researchers were doing exactly that, extracting tissue and letting it live in a dish for a short time. Today, most tissue culture experiments are done using cells rather than tissues. Much of what we know about cancer, heritable diseases, and the effects of the environment on human health has been derived from studies of cultured cells. | |
| − | + | ||
| − | + | What types of cells do people study, and from where do they come? Cells acquired directly from animal tissue are called primary cells. They are difficult to culture, largely because primary cells in this context divide only a limited number of times. This limitation on the lifespan of cultured primary cells, called the Hayflick limit, is a problem because it requires a researcher to repeatedly remove tissues from animals to complete a study. Cell isolation processes can be quite labor-intensive, and also can complicate data analysis due to inherent animal-to-animal variation. To get around this problem, researchers study cells that are immortal, which means the cells are able to divide indefinitely. Though using immortal cells is preferable for many reasons, some inherent cell-to-cell variation still exists in such populations and the genetic changes that cause immortality may affect experimental outcomes. | |
| − | + | ||
| − | + | The art of tissue culture lies in the ability to create conditions that are similar to what a cell would experience in an animal, namely 37°C and neutral pH. Blood nourishes the cells in an animal, and blood components are therfore used to feed cells in culture. Serum, the cell-free (and clotting-factor free) component of blood, contains many of the factors necessary to support the growth of cells outside the animal. Consequently, serum is frequently added to tissue culture medium, although serum-free media (also called chemically defined media) exist and support some types of cultured cells. | |
| − | + | Cultured mammalian cells must grow in a germ-free environment and researchers using tissue culture must be skilled in sterile or asceptic technique. Bacteria double very quickly relative to mammalian cells. An average mammalian cell doubles about once per day whereas many common bacteria can double every 20 minutes under optimal conditions. Consequently, if you were to co-culture 100 mammalian cells and 1 bacteria together in a flask, within 24 hours you would have ~200 unhappy mammalian cells, and about 100 million happy bacteria! Needless to say, you would not find it very useful to continue to study the behavior of your mammalian cells under these conditions. | |
| − | + | ||
| − | + | Today you will learn and practice asceptic technique to collect and count cells that were previously cultured by the teaching faculty. You will use the cells you collect to seed (or culture) cells for Western blot analysis. To ensure you are well-prepared for your work in the tissue culture room, your instructor will deliver the prelab information to you during a demo at the tissue culture hood. Because the tissue culture room is not large enough to accommodate the entire class at once, we will be split into two groups today...and all subsequent days that involve tissue culture. Half of the class will begin in tissue culture with Part 2 and the other half of the class will begin by learning more about the cell lines we are using with Part 3. Your instructor will designate the groups. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
==Protocols== | ==Protocols== | ||
| + | ===Part 1: BE Communication Lab workshop=== | ||
| + | One of the major assignments of this module is the Journal Club presentation. Conveying your science orally to an audience is an important skill that requires practice, practice, and practice. To prepare you for this task, today we will be joined by Dr. Diana Chien and Dr. Vivian Siegel to discuss slide design and how to best deliver the information to your audience. | ||
| − | + | Please find the [[Media:20.109 Workshop 3 - Journal Club 2016.pdf| Slide Presentation]] from the BE Communication Lab to assist you with your assignments. | |
| − | + | ||
| − | + | ===Part 2: Prepare cells for Western blot analysis=== | |
| − | + | '''We will begin with prelab - a discussion concerning the media and a demo of sterile technique and how to use the tissue culture hoods.''' | |
| − | + | Each team will receive two T25 culture flasks: one with M059K (wild-type) cells, and one with M059J (DNA-PK mutant) cells. You will enzymatically detach these adherent cells, count them, and plate them at a specific seeding density in a 60 mm culture dish. Next time, after these cells have doubled a couple of times, you will lyse them to make cell free extracts and begin your first assay of the module. For this reason, it is important that you take care to use sterile technique today. | |
| − | == | + | <font color=red>'''It is essential that you do not mix up or cross-contaminate the M059K and M059J cell lines! We suggest that one partner is responsible for each flask, and that partners do not share Pasteur pipets or other equipment.'''</font color=red> |
| − | + | ====Collecting cells==== | |
| + | #The tissue culture (TC) hood is partly set up for you. Finish preparing your hood according to the demo, first bringing in any remaining equipment you will need, then picking up the pre-warmed reagents from the water bath, and finally obtaining your cells. Don't forget to spray everything down with 70% ethanol! | ||
| + | #*One of the greatest sources for TC contamination is moving materials in and out of the hood because this disturbs the air flow that maintains a sterile environment inside the hood. Think about what you will need during your experiment to avoid moving your arms in and out of the hood while you are handling your cells. | ||
| + | #Examine your cell cultures as you remove the flasks from the incubator. | ||
| + | #*Look first at the color and clarity of the media. Fresh media is reddish-orange in color and if the media in your flask is yellow or cloudy, it could mean that the cells are overgrown, contaminated, or starved for CO<sub>2</sub>. | ||
| + | #*Next, look at the cells using the inverted microscope. Note their shape, arrangement, and how densely the cells cover the surface of the flask. | ||
| + | #After you look at your cells, take the flask to your tissue culture hood to begin the seeding procedure. | ||
| + | #Aspirate the media from the cells using a sterile Pasteur pipet. | ||
| + | #Wash the cells by adding 3 mL PBS using a 5 mL pipet. Slightly tip the flask back and forth to rinse the cells then aspirate the PBS with a '''fresh''' Pasteur pipet. | ||
| + | #To dislodge the cells from the flask, you will add trypsin, a proteolytic enzyme. | ||
| + | #*With a 2 mL pipet, add 1 mL of trypsin to the flask. | ||
| + | #*2 mL pipets are tricky! They fill up quickly. '''Be careful not to pull up the liquid too quickly or it will go all the way up your pipet into the pipet-aid!''' If this happens, please alert the teaching faculty rather than returning the pipet-aid to the rack. | ||
| + | #Tip the flask in each direction to distribute the trypsin evenly then incubate the cells at 37°C for 2 minutes using a timer. | ||
| + | #*While you are waiting, label your 60 mm dish. Include your group color and today’s date. Then add the cell strain name and the initials of the partner who prepared those particular cells. | ||
| + | #*Finally, this is a great time to clear out your trash! | ||
| + | #Retrieve your flasks from the incubator and firmly tap the bottom 5x to dislodge the cells. | ||
| + | #*Check your cells using the microscope to ensure they are dislodged. The should appear round and move freely. | ||
| + | #*If your cells are not detached from the flask, incubate at 37°C for an additional minute. | ||
| + | #When your cells are dislodged, move your flask back into the TC hood and add 3 mL of media to the cells then pipet the liquid up and down (“triturate”) to break up cells that are clumped together and suspend them in the liquid. | ||
| + | #*'''Note: do not take up or release all the liquid, in order to avoid bubbles.''' | ||
| + | #Transfer 90 μL of your cell suspension into a labeled eppendorf tube. | ||
| + | #*Be sure to cap your conical tube and eppendorf tube after you transfer your cells. | ||
| − | + | ====Counting and seeding cells==== | |
| + | #Remove the eppendorf with your cells from the TC hood. | ||
| + | #Add 10 μL of Trypan blue to the eppendorf tube and pipet up and down to mix. | ||
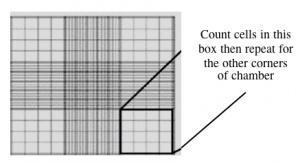
| + | #With a new pipet tip, transfer 10 μL of the cell/dye suspension to a hemocytometer, as shown to you by the teaching faculty. Keep M059K on 'top' and M059J on the 'bottom', so you don't forget which is which. [[Image:Be109cellcounting.jpg|thumb|right|300px|'''Counting cells using a hemocytometer.''']] | ||
| + | #Count the cells within two diagonal corners, as shown to you by the teaching faculty. If the values are within 10% of each other, continue. If they are more different than that, count the other two corners. Be sure to record all of your raw data. | ||
| + | #*To obtain your actual cell count, take the average of either the two or the four values. | ||
| + | #*The hemocytometer has an etched grid of nine large squares. The concentration of cells in a sample can be determined by counting the cells that fall within one such square and then multiplying by 10,000 to determine the number of cells/mL in the solution measured. (Always remember to account for the dilution to cell stock at the end!) | ||
| + | #*Note that different squares are sub-divided into different grids. Very dense cells could be counted in the fine grids. In your case, the 4 x 4 grids and a 10x magnification will be most convenient for counting. | ||
| + | #Calculate the volume of cells you need to seed 100,000 cells in your 60 mm dish. | ||
| + | #*For example, if your concentration is 1 million (1 M) cells/mL, you would take 0.1 mL of cells and 2.9 mL of media. | ||
| + | #*Make this new cell suspension in the 60 mm plate -- first add your cells and then add fresh media to a total volume of 3 mL. | ||
| + | #*You should mix your ''original'' cell suspension by pipetting before distributing the small volume of cells to the new dish as the cells likely settled to the bottom of the conical while you were counting. | ||
| + | #Finally, tilt your 60 mm dish back and forth to distribute the cells evenly. | ||
| − | ==== | + | ====Cleaning out the TC hood==== |
| + | The next group who uses your hood should find the surfaces wiped down and free of equipment. '''Please leave the equipment that was already there.''' | ||
| + | #Aspirate any remaining cell suspensions. | ||
| + | #Dispose of all vessels that held cells in the biohazard waste box and be sure that all sharps are in the mayo jar. | ||
| + | #Remove any equipment that you transferred into the hood and return it to the appropriate location. | ||
| + | #Spray the TC hood surface with 70% ethanol and wipe with paper towels. | ||
| + | #*Be sure the paper towels are disposed of in the biohazard waste box! | ||
| + | #Empty the benchtop biohazard bucket into the biohazard waste box. | ||
| − | + | ===Part 3: Research the M059K and M059J cell lines=== | |
| − | + | In this exercise you will learn more about the cell lines you are using in this module. The wild-type strain is called M059K. Wild type is used to describe the phenotype of a typical or non-mutant form of a bacterial species or cell line. The mutant strain is called M059J and does not generate a subunit of DNA-PK, rendering the protein inactive. A great resource from which we can gather information on these cell lines is the American Tissue Type Collection (ATCC). Use the information concerning [http://www.atcc.org/products/all/CRL-2365.aspx M059K] and [http://www.atcc.org/products/all/CRL-2366.aspx M059J] to answer the following questions in your notebook: | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | #When were these fibroblast cells originally acquired? | |
| + | #What characteristic of the cell is described by the term 'fibroblast'? | ||
| + | #Do these cells float in the culture media, or stick to the culture flask? How do you know? | ||
| + | #From what organ tissue were these cells harvested? | ||
| + | #For what types of studies are these cells a useful model system? What evidence is provided to support this? | ||
| + | #How should these cells be cited in a scientific publication? | ||
| − | + | The M059J strain was first described by Allalunis-Turner ''et al.'' in a research article [http://www.rrjournal.org/doi/abs/10.2307/3578196 found here]. Use this paper to answer the following questions in your notebook: | |
| − | The | + | #Read the introduction. |
| + | #*Why might it be useful to have model cell lines that exhibit sensitivity to radiation or drugs? | ||
| + | #*The purpose of an introduction is to provide the reader with information necessary for understanding the purpose and results of the paper. Is there any additional information that would be helpful to include in this introduction? | ||
| + | #Read the results. | ||
| + | #*Review Fig. 3. In the results section, the authors state that the "SF2 values calculated for M059J and M059K cells were 0.02 and 0.64, respectively." Read the portion of the ''Colony-forming assay'' paragraph that discusses the radiation assay in the methods section. Briefly, discuss how these data were generated. What additional information would be useful if you wanted to repeat this experiment? | ||
| + | #*Review Fig. 4 and 5. What do these data suggest concerning the role of DNA-PK in dsb repair? Consider a possible mechanism by which the drugs are interacting with DNA-PK. | ||
| + | #*Review Fig. 6. Are the results for M059J and M059K what you expect? Why or why not? | ||
| + | #Read the discussion. | ||
| + | #*Why is the authors' approach to acquiring cell lines from individual tumors with differing properties preferable? | ||
| + | #*Within the discussion authors often present caveats of their research. What do the authors note about their cell lines that should be considered by the reader? | ||
| − | + | The radiosensitivity phenotype of M059J was further explored by Lees-Miller ''et al.'' in a research article [http://www.ncbi.nlm.nih.gov/pubmed/7855602 found here]. Use this paper to answer the following questions in your notebook: | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | #Read paragraphs 1-4. | |
| − | + | #*Draw a diagram that includes a DNA double-strand break (dsb), DNA-PK, Ku70, and Ku80, depicting the relationship between the molecules. Feel free to use sources outside of the paper. | |
| − | * | + | #*What is known about the function/role of DNA-PK? |
| − | + | #*What do the authors suggest is the reason for the radiosensitivity phenotype of M059J cells? | |
| − | + | #Read paragraphs 5-9. | |
| − | + | #*What experiment(s) did the authors complete to test for the presence of the DNA-PK protein? the gene that encodes DNA-PK? the activity of DNA-PK? | |
| − | + | #Read paragraphs 10-end. | |
| − | + | #*What do the authors suggest is the root cause of the M059J phenotype? Do the data support this interpretation? Why or why not? | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | * | + | |
| − | + | ||
| − | + | ||
==Reagent list== | ==Reagent list== | ||
| − | + | Glioblastoma cells from ATCC | |
| − | + | *M059K: plated at 100,000 cells per T25 flask 1 day in advance | |
| − | + | *M059J: plated at 100,000 cells per T25 flask 1 day in advance | |
Media components from Life Technologies, Inc. unless noted otherwise. | Media components from Life Technologies, Inc. unless noted otherwise. | ||
| Line 126: | Line 134: | ||
**100X antibiotic solution from Cellgro | **100X antibiotic solution from Cellgro | ||
***10,000 U/mL Penicillin | ***10,000 U/mL Penicillin | ||
| − | ***10,000 | + | ***10,000 U/mL Streptomycin |
**100X non-essential amino acids (NEAA) | **100X non-essential amino acids (NEAA) | ||
***varying amounts of seven amino acids | ***varying amounts of seven amino acids | ||
| Line 133: | Line 141: | ||
*0.25% trypsin/0.91 mM EDTA | *0.25% trypsin/0.91 mM EDTA | ||
| + | |||
| + | *Incubator maintains 37°C, 5% CO2 and 95% relative humidity | ||
==Navigation links== | ==Navigation links== | ||
Latest revision as of 17:34, 19 September 2016
Contents
Introduction
To be confident of your findings related to DNA repair, you will first validate the cell lines you are using to assess the effect of DNA damage and drugs on NHEJ. Just as it was important to confirm the template you used for your mutagenesis reaction in Mod 1, it is important to confirm the cells you are using are indeed mutants in the protein you are assessing in this module. You will do this by completing a Western blot. A Western blot allows researchers to detect specific proteins within a mixed sample, such as cell lysate. We will use this protein analysis technique to assess DNA-PK levels in the wild-type M059K and DNA-PK mutant M059J cell lines.
One important objective for this experimental module is for you to learn how to maintain cultured mammalian cell lines, or to perform tissue culture. Today you will jump right into this task as you seed M059K and M059J cells for your Western protein assay. These human fibroblast cell lines were originally acquired from a malignant glioblastoma.Tissue culture was developed ~100 years ago as a means to study mammalian biology, and since that time we have learned a tremendous amount by observing the behavior of mammalian cells maintained in the laboratory. The term tissue culture was originally coined because researchers were doing exactly that, extracting tissue and letting it live in a dish for a short time. Today, most tissue culture experiments are done using cells rather than tissues. Much of what we know about cancer, heritable diseases, and the effects of the environment on human health has been derived from studies of cultured cells.
What types of cells do people study, and from where do they come? Cells acquired directly from animal tissue are called primary cells. They are difficult to culture, largely because primary cells in this context divide only a limited number of times. This limitation on the lifespan of cultured primary cells, called the Hayflick limit, is a problem because it requires a researcher to repeatedly remove tissues from animals to complete a study. Cell isolation processes can be quite labor-intensive, and also can complicate data analysis due to inherent animal-to-animal variation. To get around this problem, researchers study cells that are immortal, which means the cells are able to divide indefinitely. Though using immortal cells is preferable for many reasons, some inherent cell-to-cell variation still exists in such populations and the genetic changes that cause immortality may affect experimental outcomes.
The art of tissue culture lies in the ability to create conditions that are similar to what a cell would experience in an animal, namely 37°C and neutral pH. Blood nourishes the cells in an animal, and blood components are therfore used to feed cells in culture. Serum, the cell-free (and clotting-factor free) component of blood, contains many of the factors necessary to support the growth of cells outside the animal. Consequently, serum is frequently added to tissue culture medium, although serum-free media (also called chemically defined media) exist and support some types of cultured cells.
Cultured mammalian cells must grow in a germ-free environment and researchers using tissue culture must be skilled in sterile or asceptic technique. Bacteria double very quickly relative to mammalian cells. An average mammalian cell doubles about once per day whereas many common bacteria can double every 20 minutes under optimal conditions. Consequently, if you were to co-culture 100 mammalian cells and 1 bacteria together in a flask, within 24 hours you would have ~200 unhappy mammalian cells, and about 100 million happy bacteria! Needless to say, you would not find it very useful to continue to study the behavior of your mammalian cells under these conditions.
Today you will learn and practice asceptic technique to collect and count cells that were previously cultured by the teaching faculty. You will use the cells you collect to seed (or culture) cells for Western blot analysis. To ensure you are well-prepared for your work in the tissue culture room, your instructor will deliver the prelab information to you during a demo at the tissue culture hood. Because the tissue culture room is not large enough to accommodate the entire class at once, we will be split into two groups today...and all subsequent days that involve tissue culture. Half of the class will begin in tissue culture with Part 2 and the other half of the class will begin by learning more about the cell lines we are using with Part 3. Your instructor will designate the groups.
Protocols
Part 1: BE Communication Lab workshop
One of the major assignments of this module is the Journal Club presentation. Conveying your science orally to an audience is an important skill that requires practice, practice, and practice. To prepare you for this task, today we will be joined by Dr. Diana Chien and Dr. Vivian Siegel to discuss slide design and how to best deliver the information to your audience.
Please find the Slide Presentation from the BE Communication Lab to assist you with your assignments.
Part 2: Prepare cells for Western blot analysis
We will begin with prelab - a discussion concerning the media and a demo of sterile technique and how to use the tissue culture hoods.
Each team will receive two T25 culture flasks: one with M059K (wild-type) cells, and one with M059J (DNA-PK mutant) cells. You will enzymatically detach these adherent cells, count them, and plate them at a specific seeding density in a 60 mm culture dish. Next time, after these cells have doubled a couple of times, you will lyse them to make cell free extracts and begin your first assay of the module. For this reason, it is important that you take care to use sterile technique today.
It is essential that you do not mix up or cross-contaminate the M059K and M059J cell lines! We suggest that one partner is responsible for each flask, and that partners do not share Pasteur pipets or other equipment.
Collecting cells
- The tissue culture (TC) hood is partly set up for you. Finish preparing your hood according to the demo, first bringing in any remaining equipment you will need, then picking up the pre-warmed reagents from the water bath, and finally obtaining your cells. Don't forget to spray everything down with 70% ethanol!
- One of the greatest sources for TC contamination is moving materials in and out of the hood because this disturbs the air flow that maintains a sterile environment inside the hood. Think about what you will need during your experiment to avoid moving your arms in and out of the hood while you are handling your cells.
- Examine your cell cultures as you remove the flasks from the incubator.
- Look first at the color and clarity of the media. Fresh media is reddish-orange in color and if the media in your flask is yellow or cloudy, it could mean that the cells are overgrown, contaminated, or starved for CO2.
- Next, look at the cells using the inverted microscope. Note their shape, arrangement, and how densely the cells cover the surface of the flask.
- After you look at your cells, take the flask to your tissue culture hood to begin the seeding procedure.
- Aspirate the media from the cells using a sterile Pasteur pipet.
- Wash the cells by adding 3 mL PBS using a 5 mL pipet. Slightly tip the flask back and forth to rinse the cells then aspirate the PBS with a fresh Pasteur pipet.
- To dislodge the cells from the flask, you will add trypsin, a proteolytic enzyme.
- With a 2 mL pipet, add 1 mL of trypsin to the flask.
- 2 mL pipets are tricky! They fill up quickly. Be careful not to pull up the liquid too quickly or it will go all the way up your pipet into the pipet-aid! If this happens, please alert the teaching faculty rather than returning the pipet-aid to the rack.
- Tip the flask in each direction to distribute the trypsin evenly then incubate the cells at 37°C for 2 minutes using a timer.
- While you are waiting, label your 60 mm dish. Include your group color and today’s date. Then add the cell strain name and the initials of the partner who prepared those particular cells.
- Finally, this is a great time to clear out your trash!
- Retrieve your flasks from the incubator and firmly tap the bottom 5x to dislodge the cells.
- Check your cells using the microscope to ensure they are dislodged. The should appear round and move freely.
- If your cells are not detached from the flask, incubate at 37°C for an additional minute.
- When your cells are dislodged, move your flask back into the TC hood and add 3 mL of media to the cells then pipet the liquid up and down (“triturate”) to break up cells that are clumped together and suspend them in the liquid.
- Note: do not take up or release all the liquid, in order to avoid bubbles.
- Transfer 90 μL of your cell suspension into a labeled eppendorf tube.
- Be sure to cap your conical tube and eppendorf tube after you transfer your cells.
Counting and seeding cells
- Remove the eppendorf with your cells from the TC hood.
- Add 10 μL of Trypan blue to the eppendorf tube and pipet up and down to mix.
- With a new pipet tip, transfer 10 μL of the cell/dye suspension to a hemocytometer, as shown to you by the teaching faculty. Keep M059K on 'top' and M059J on the 'bottom', so you don't forget which is which.
- Count the cells within two diagonal corners, as shown to you by the teaching faculty. If the values are within 10% of each other, continue. If they are more different than that, count the other two corners. Be sure to record all of your raw data.
- To obtain your actual cell count, take the average of either the two or the four values.
- The hemocytometer has an etched grid of nine large squares. The concentration of cells in a sample can be determined by counting the cells that fall within one such square and then multiplying by 10,000 to determine the number of cells/mL in the solution measured. (Always remember to account for the dilution to cell stock at the end!)
- Note that different squares are sub-divided into different grids. Very dense cells could be counted in the fine grids. In your case, the 4 x 4 grids and a 10x magnification will be most convenient for counting.
- Calculate the volume of cells you need to seed 100,000 cells in your 60 mm dish.
- For example, if your concentration is 1 million (1 M) cells/mL, you would take 0.1 mL of cells and 2.9 mL of media.
- Make this new cell suspension in the 60 mm plate -- first add your cells and then add fresh media to a total volume of 3 mL.
- You should mix your original cell suspension by pipetting before distributing the small volume of cells to the new dish as the cells likely settled to the bottom of the conical while you were counting.
- Finally, tilt your 60 mm dish back and forth to distribute the cells evenly.
Cleaning out the TC hood
The next group who uses your hood should find the surfaces wiped down and free of equipment. Please leave the equipment that was already there.
- Aspirate any remaining cell suspensions.
- Dispose of all vessels that held cells in the biohazard waste box and be sure that all sharps are in the mayo jar.
- Remove any equipment that you transferred into the hood and return it to the appropriate location.
- Spray the TC hood surface with 70% ethanol and wipe with paper towels.
- Be sure the paper towels are disposed of in the biohazard waste box!
- Empty the benchtop biohazard bucket into the biohazard waste box.
Part 3: Research the M059K and M059J cell lines
In this exercise you will learn more about the cell lines you are using in this module. The wild-type strain is called M059K. Wild type is used to describe the phenotype of a typical or non-mutant form of a bacterial species or cell line. The mutant strain is called M059J and does not generate a subunit of DNA-PK, rendering the protein inactive. A great resource from which we can gather information on these cell lines is the American Tissue Type Collection (ATCC). Use the information concerning M059K and M059J to answer the following questions in your notebook:
- When were these fibroblast cells originally acquired?
- What characteristic of the cell is described by the term 'fibroblast'?
- Do these cells float in the culture media, or stick to the culture flask? How do you know?
- From what organ tissue were these cells harvested?
- For what types of studies are these cells a useful model system? What evidence is provided to support this?
- How should these cells be cited in a scientific publication?
The M059J strain was first described by Allalunis-Turner et al. in a research article found here. Use this paper to answer the following questions in your notebook:
- Read the introduction.
- Why might it be useful to have model cell lines that exhibit sensitivity to radiation or drugs?
- The purpose of an introduction is to provide the reader with information necessary for understanding the purpose and results of the paper. Is there any additional information that would be helpful to include in this introduction?
- Read the results.
- Review Fig. 3. In the results section, the authors state that the "SF2 values calculated for M059J and M059K cells were 0.02 and 0.64, respectively." Read the portion of the Colony-forming assay paragraph that discusses the radiation assay in the methods section. Briefly, discuss how these data were generated. What additional information would be useful if you wanted to repeat this experiment?
- Review Fig. 4 and 5. What do these data suggest concerning the role of DNA-PK in dsb repair? Consider a possible mechanism by which the drugs are interacting with DNA-PK.
- Review Fig. 6. Are the results for M059J and M059K what you expect? Why or why not?
- Read the discussion.
- Why is the authors' approach to acquiring cell lines from individual tumors with differing properties preferable?
- Within the discussion authors often present caveats of their research. What do the authors note about their cell lines that should be considered by the reader?
The radiosensitivity phenotype of M059J was further explored by Lees-Miller et al. in a research article found here. Use this paper to answer the following questions in your notebook:
- Read paragraphs 1-4.
- Draw a diagram that includes a DNA double-strand break (dsb), DNA-PK, Ku70, and Ku80, depicting the relationship between the molecules. Feel free to use sources outside of the paper.
- What is known about the function/role of DNA-PK?
- What do the authors suggest is the reason for the radiosensitivity phenotype of M059J cells?
- Read paragraphs 5-9.
- What experiment(s) did the authors complete to test for the presence of the DNA-PK protein? the gene that encodes DNA-PK? the activity of DNA-PK?
- Read paragraphs 10-end.
- What do the authors suggest is the root cause of the M059J phenotype? Do the data support this interpretation? Why or why not?
Reagent list
Glioblastoma cells from ATCC
- M059K: plated at 100,000 cells per T25 flask 1 day in advance
- M059J: plated at 100,000 cells per T25 flask 1 day in advance
Media components from Life Technologies, Inc. unless noted otherwise.
- M059J/K cell media
- 1:1 Dulbecco's Modified Eagle's Medium (DMEM) : Ham's F12 medium
- 2.5 mM L-Glutamine
- 15 mM HEPES
- 0.5 mM sodium pyruvate
- 1.2 g/L sodium bicarbonate
- 10% fetal bovine serum (FBS)
- 100X antibiotic solution from Cellgro
- 10,000 U/mL Penicillin
- 10,000 U/mL Streptomycin
- 100X non-essential amino acids (NEAA)
- varying amounts of seven amino acids
- 1:1 Dulbecco's Modified Eagle's Medium (DMEM) : Ham's F12 medium
- Dulbecco's phosphate-buffered saline (D-PBS)
- 0.25% trypsin/0.91 mM EDTA
- Incubator maintains 37°C, 5% CO2 and 95% relative humidity
Next day: Begin Western protein analysis