20.109(F23):M2D5
Contents
Introduction
Interactions between low molecular weight ligands, or small molecules, and proteins have been shown to increase the thermostability of proteins. This means that proteins bound to ligand are able to maintain tertiary structure, or resist denaturation, at higher temperatures than unbound proteins. Today we will use differential scanning fluorimetry (DSF) to examine the putative PfFKBP35 small molecule binders.
DSF is a method used to identify low molecular weight ligands that bind and stabilize a protein of interest. In this assay, protein denaturation is measured via a fluorescent dye that has an affinity for hydrophobic regions. When the protein is folded the hydrophobic pockets are inaccessible to the dye and the fluorescent signal is quenched by water in the solution. As the protein unfolds, the dye interacts with the hydrophobic regions and emits a fluorescent signal that can be detected.
When a protein is bound to a ligand, the stability can be increased such that the temperature at which the protein denatures is increased. In the DSF assay, this is measured as a shift in the Tm, or melting temperature; which is defined as the temperature at which 50% of the protein is unfolded. This value represents the midpoint of the transition from structured (folded) to denatured (unfolded).
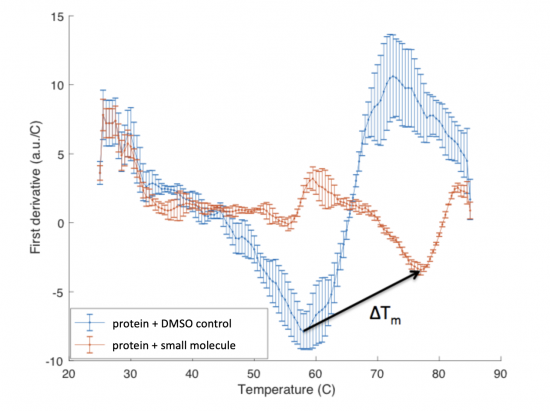
The ΔTm is the difference between the Tm of the unbound protein sample, or protein sample without added ligand, and the bound protein sample, protein sample with added ligand. If the tested ligand binds the protein of interest, the ΔTm can be observed as a shift in the plotted DSF data. For example, the data below show results of a pilot experiment completed in preparation for this module. In this graph the Tm of FKBP12 (blue curve) is ~50 °C. With the addition of rapamycin (red curve) the Tm is shifted to ~78 °C resulting in a ΔTm of ~20 degrees.
Protocols
Part 1: Complete DSF experiment
The protein you purified in the previous laboratory session was pooled and prepared for use in the DSF assay. Specifically, the purified protein pool was diluted in 1X TBS to a final concentration of 0.25 mg/mL. For this experiment you will use a serial dilution of two of the compounds you chose to generate dose response curves. Please find the compound assignments in the table below:
| Team | Compound #1 | Compound #2 |
| TR Yellow | FK141 | FK502 |
| TR Green | FK296 | FK502 |
| TR Blue | FK141 | FK189 |
| TR Purple | FK189 | FK502 |
| WF Green | FK189 | FK296 |
| WF Blue | FK141 | FK296 |
Please note that in some cases you may be testing small molecules different than those selected in the previous laboratory section. To ensure that all small molecules were included and to ensure that replicates were completed for all small molecules, some changes were made to enhance the class data. Everyone will have access to all of the class data.
- Obtain two aliquots of the prepared PfFKBP35 protein from the front laboratory bench.
- Each aliquot will be used to test one compound. To divide the work, each laboratory partner will be responsible for completing the below steps with one of the compounds.
- Add 7.3 μL of Sypro Orange to the prepared PfFKBP35 protein and mix gently by pipetting.
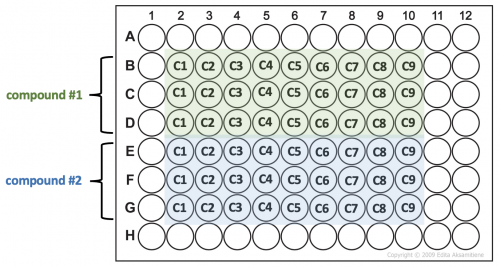
- For each compound, Label 9 microcentrifuge tubes ('C1' through 'C9') to reflect the concentrations of the compounds that will be used in your experiment.
- Add the specified volume of your protein sample to each tube:
- To the tube labeled 'C1' add 133 μL of protein.
- To the tubes labeled 'C2' through 'C9' add 70 μL of protein.
- Tube 'C6' is an exception - add 84 μL of protein instead.
- You will prepare a serial dilution using the protein solution in the tubes as the diluent to reduce waste of the compounds. To ensure that the protocol for the dilution series is clear, first draw a schematic of the dilution scheme as written in Steps #6 and #7 then confirm with the Instructor that you are ready to proceed with the dilution steps.
- To prepare the sample with the highest concentration of compound, add 7 μL of the compound stock to the tube labeled 'C1'.
- Carefully bring your 'C1' tube to the front bench and your Instructor will add the compound for you from the stock.
- Next, complete serial dilutions to prepare the remainder of your protein samples with decreasing concentrations of compound.
- Pipet 70 μL from tube 'C1' into tube 'C2', then mix by gently pipetting. Do not vortex samples!
- This will result in 1:2 dilution of the compound from tube 'C1' to tube 'C2'.
- Pipet 70 μL from tube 'C2' into tube 'C3', then mix by gently pipetting.
- Pipet 70 μL from tube 'C3' into tube 'C4', then mix by gently pipetting.
- Pipet 70 μL from tube 'C4' into tube 'C5', then mix by gently pipetting.
- Pipet 56 μL from tube 'C5' into tube 'C6', then mix by gently pipetting. Note the difference in volume pipetted here.
- Pipet 70 μL from tube 'C6' into tube 'C7', then mix by gently pipetting.
- Pipet 70 μL from tube 'C7' into tube 'C8', then mix by gently pipetting.
- Do not pipet into tube 'C9' as this is the no compound control!
- Pipet 70 μL from tube 'C1' into tube 'C2', then mix by gently pipetting. Do not vortex samples!
- Transfer 20 μL aliquots from each tube into three wells of a 96-well plate according to the plate map below.
- Alert your Instructor when your plate is loaded as the samples will be measured in the BioMicro Center.
In your laboratory notebook, complete the following:
- Calculate the final concentration of protein used in the DSF experiment.
- Concentration of prepared protein = 0.25 mg/mL
- Calculate the final concentration of small molecule compound used in each of the prepared samples ('C1' through 'C9').
- Concentration of small molecule stocks = 1 mM
Reagents list
- PfFKBP35 (concentration = 0.25 mg/mL)
- Sypro Orange dye (from Thermo Fisher)
- small molecule compounds (1 mM) (from Chembridge)
Next day: Complete data analysis of functional assay results