Difference between revisions of "20.109(F09): Mod 2 Day 2 Measuring system performance"
| Line 17: | Line 17: | ||
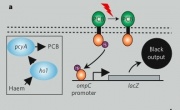
This figure is more detailed but harder to decipher without prior understanding of several aspects of the system. For example, light at 660nm is detected by the input-sensing device, turning it off. Light-sensing requires the combined action of two sets of proteins: Cph8, itself a fusion of a light-sensing protein called Cph1 that comes from an algae attached to a transmembrane signaling protein called EnvZ, and phycobilin producing proteins that generate accessory pigments needed for the light-sensing protein to work. The input sensing device generates a signal within the cell’s osmoregulation pathway (OmpR is the signal carrier), changing the activity of an OmpR regulated promoter that is directing transcription of the output: lacZ….whew! The black boxes were a whole lot easier. | This figure is more detailed but harder to decipher without prior understanding of several aspects of the system. For example, light at 660nm is detected by the input-sensing device, turning it off. Light-sensing requires the combined action of two sets of proteins: Cph8, itself a fusion of a light-sensing protein called Cph1 that comes from an algae attached to a transmembrane signaling protein called EnvZ, and phycobilin producing proteins that generate accessory pigments needed for the light-sensing protein to work. The input sensing device generates a signal within the cell’s osmoregulation pathway (OmpR is the signal carrier), changing the activity of an OmpR regulated promoter that is directing transcription of the output: lacZ….whew! The black boxes were a whole lot easier. | ||
| − | Today you will assess the system, both in terms of enzymatic activity measurements as well as the | + | Today you will assess the system, both in terms of enzymatic activity measurements as well as a resulting bacterial photograph. Next time we'll start to "tune" the system in order to expand the dynamic range of the system. |
==Protocols== | ==Protocols== | ||
| Line 27: | Line 27: | ||
===Part 2: Black and white photography=== | ===Part 2: Black and white photography=== | ||
| − | Retrieve the Petri dishes you set up last time and compare the appearance of the light and dark grown samples. Because the dark grown cells were in a completely dark box, the difference between the two plates is the greatest contrast you can expect in your bacterial photographs. | + | Retrieve the Petri dishes you set up last time and compare the appearance of the light and dark grown samples. Because the dark grown cells were in a completely dark box, the difference between the two plates is the greatest contrast you can expect in your bacterial photographs. You should photograph these petri dishes since the result will be included in the [[20.109(F09): System engineering research article| final research article]] you write for this module. |
| − | Next decide what image you would like to photograph. Generate a computer file with this image and print it to a transparency. Transparencies will be available in the lab for you to use as masks, taping them to the | + | Next decide what image you would like to photograph. Generate a computer file with this image and print it to a transparency. Transparencies will be available in the lab for you to use as masks, taping them to the saran covering the Petri dish before incubating. As you choose an image, remember that the goal is to have each cell growing distinctly in the light or dark. Remember that light can bounce around edges and may blur the resulting image if the black and white are highly intermingled. In general, it’s better to have a dark background and a light image rather than the other way around. To darken the dark parts of your photo, you might want to print it on two transparencies and use them both to mask your Petri dish. |
| + | |||
| + | Media containing S-gal is available for you to supplement with antibiotics and cells as you did [[20.109(F09): Mod 2 Day 1 Testing an engineered biological system| last time.]] Place your bacterial photograph in the 37° incubator and when the class is ready, we'll turn on the red-lamp to expose the "coliroids" until next time. | ||
===Part 3: Instruction on Oral Presentations=== | ===Part 3: Instruction on Oral Presentations=== | ||
| Line 37: | Line 39: | ||
==For Next Time== | ==For Next Time== | ||
| + | Calculate the units associated with the | ||
==Reagents== | ==Reagents== | ||
Revision as of 18:59, 25 August 2009
Contents
Measuring System Performance
Introduction
Operation of System's Devices
The bacterial photography system we are studying modifies a natural signaling pathway in E. coli so that light striking the cell can be converted to a detectable output (β-galactosidase enzyme that works on a chromogenic substance).
2CS K+ and P+ activities
Black box abstraction
A “black box” depiction of the system looks like:
Some of the details within each box are:
This figure is more detailed but harder to decipher without prior understanding of several aspects of the system. For example, light at 660nm is detected by the input-sensing device, turning it off. Light-sensing requires the combined action of two sets of proteins: Cph8, itself a fusion of a light-sensing protein called Cph1 that comes from an algae attached to a transmembrane signaling protein called EnvZ, and phycobilin producing proteins that generate accessory pigments needed for the light-sensing protein to work. The input sensing device generates a signal within the cell’s osmoregulation pathway (OmpR is the signal carrier), changing the activity of an OmpR regulated promoter that is directing transcription of the output: lacZ….whew! The black boxes were a whole lot easier.
Today you will assess the system, both in terms of enzymatic activity measurements as well as a resulting bacterial photograph. Next time we'll start to "tune" the system in order to expand the dynamic range of the system.
Protocols
Be sure to retrieve your light and dark plates from the incubator and photograph them to illustrate the maximal contrast associated with the starting system.
Part 1: β-galactosidase assay
You should perform assays on your overnight liquid cultures that were grown in the dark and the light. Refer back to the protocol from day 1. Activity calculations for these samples will be part of your assignment for next time.
Part 2: Black and white photography
Retrieve the Petri dishes you set up last time and compare the appearance of the light and dark grown samples. Because the dark grown cells were in a completely dark box, the difference between the two plates is the greatest contrast you can expect in your bacterial photographs. You should photograph these petri dishes since the result will be included in the final research article you write for this module.
Next decide what image you would like to photograph. Generate a computer file with this image and print it to a transparency. Transparencies will be available in the lab for you to use as masks, taping them to the saran covering the Petri dish before incubating. As you choose an image, remember that the goal is to have each cell growing distinctly in the light or dark. Remember that light can bounce around edges and may blur the resulting image if the black and white are highly intermingled. In general, it’s better to have a dark background and a light image rather than the other way around. To darken the dark parts of your photo, you might want to print it on two transparencies and use them both to mask your Petri dish.
Media containing S-gal is available for you to supplement with antibiotics and cells as you did last time. Place your bacterial photograph in the 37° incubator and when the class is ready, we'll turn on the red-lamp to expose the "coliroids" until next time.
Part 3: Instruction on Oral Presentations
Over the course of the term, everyone taking 20.109 will be making two presentations to the class. One will be a presentation of a primary journal article related to either 2 component signaling systems or synthetic biology. The other presentation will be of a research idea. Today our wonderful coaches from the Writing program will come to lab to give a talk about giving talks. This presentation should be a welcomed chance to learn what's expected when you speak (in 20.109 and elsewhere) and to gather some great tips for giving polished and interesting seminars. You can follow up this instruction by re-reading the oral presentation guidelines we've assembled for this subject.
DONE!
For Next Time
Calculate the units associated with the
==Reagents==