20.109(S09):Prepare expression system (Day4)
Introduction
update schemes to reflect what is being done by students vs. instructors this semester
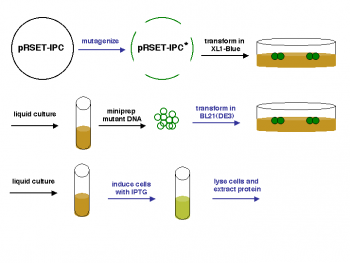
Now that we have prepared DNA encoding your mutant inverse pericams, we would like to actually produce the physical proteins. Last time you were here, you transformed competent bacteria (called XL1-Blue) with mutagenized DNA prepared from a template plasmid prepared. Successfully transformed bacteria grew into colonies on amipicillin-containing plates, and yesterday your oh-so devoted teaching staff picked two colonies per mutant to grow in liquid culture. The XL1-Blue cell line, although it now carries the inverse pericam DNA, cannot produce the inverse pericam protein. Thus, today you will extract DNA from the XL1-Blue cells, prepare it for analysis, and transform your IPC mutant plasmids into a new bacterial system that can produce the protein directly.
The bacterial expression vector we are using (pRSET) contains the bacteriophage T7 promoter. This promoter is active only in the presence of T7 RNA polymerase (T7RNAP), an enzyme that therefore must be expressed by the bacterial strain used to make the protein of interest. We will use the BL21(DE3)pLysS strain, which has the following genotype: F-, ompT hsdSB (rB- mB-) gal dcm (DE3) pLysS (CamR). In BL21(DE3), T7RNAP is associated with a lac construct, and its expression is under the control of the lacUV5 promoter. Due to the action of the lac repressor (lacI gene), the polymerase will not be produced except in the presence of lactose or a small-molecule lactose analogue such as IPTG (isopropyl β-D-thiogalactoside). To further reduce ‘leaky’ expression of the protein of interest (in our case, inverse pericam), the pLysS version of BL21(DE3) contains T7 lysozyme, which also inhibits basal transcription of T7RNAP. This gene is retained by Chloramphenicol selection, while the pRSET plasmid itself (and thus inverse pericam) is retained by Ampicillin selection - as you learned last time.
To isolate the inverse-pericam-containing pRSET plasmid from the overnight cultures, you will perform what is commonly called a “mini-prep.” This term distinguishes the procedure from a “maxi-” or “large scale-prep” which involves a larger volume of cells and additional steps of purification. The overall goal of each “prep” is the same--to separate the plasmid DNA from the chromosomal DNA and cellular debris, allowing the plasmid DNA to be studied further. In the traditional mini-prep protocol, the media is removed from the cells by centrifugation. The cells are resuspended in “Solution I” which contains Tris to buffer the cells and EDTA to bind divalent cations in the lipid bilayer, thereby weakening the cell envelope. A solution of sodium hydroxide and SDS is then added. The base denatures the cell’s DNA, both chromosomal and plasmid, while the detergent dissolves the cellular proteins and lipids. The pH of the solution is returned to neutral by the acetic acid and potassium acetate in “Solution III.” At neutral pH the SDS precipitates from solution, carrying with it the dissolved proteins and lipids. In addition, the DNA strands renature at neutral pH. The chromosomal DNA, which is much longer than the plasmid DNA, renatures as a tangle that gets trapped in the SDS precipitate. The plasmid DNA renatures normally and stays in solution, effectively separating plasmid DNA from the chromosomal DNA and the proteins and lipids of the cell.
Once you have the plasmid DNA isolated, you can prepare it for sequencing and gel analysis, as well use it immediately for transformation. In order to transform BL21(DE3) cells with your mutant IPC plasmids, you will first have to make the cells competent, i.e., able to efficiently take up foreign DNA. With the XL1-Blue strain, we used commercially available competent cells that did not need further treatment prior to DNA addition. Today, you will make chemically competent cells using calcium chloride, then incubate them with plasmid DNA and heat shock them as before prior to plating. Tomorrow, the teaching staff will pick colonies and set up liquid overnight cultures from your transformed cells. Next time, you will add IPTG to these liquid cultures to induce expression of your mutant proteins, which you will then isolate and characterize. Much of this process is summarized in the figure at left.